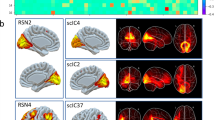
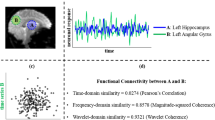
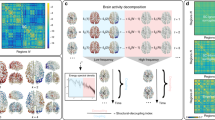
Abstract
Brain atlases are fundamental to understanding the topographic organization of the human brain, yet many contemporary human atlases cover only the cerebral cortex, leaving the subcortex a terra incognita. We use functional MRI (fMRI) to map the complex topographic organization of the human subcortex, revealing large-scale connectivity gradients and new areal boundaries. We unveil four scales of subcortical organization that recapitulate well-known anatomical nuclei at the coarsest scale and delineate 27 new bilateral regions at the finest. Ultrahigh field strength fMRI corroborates and extends this organizational structure, enabling the delineation of finer subdivisions of the hippocampus and the amygdala, while task-evoked fMRI reveals a subtle subcortical reorganization in response to changing cognitive demands. A new subcortical atlas is delineated, personalized to represent individual differences and used to uncover reproducible brain–behavior relationships. Linking cortical networks to subcortical regions recapitulates a task-positive to task-negative axis. This new atlas enables holistic connectome mapping and characterization of cortico–subcortical connectivity.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
Neuroimaging data analyses were undertaken using publicly available human neuroimaging datasets acquired and maintained by the HCP18. These datasets are available for download to anyone agreeing to the Open Access Data Use Terms (https://db.humanconnectome.org/). Access to family structure data and several behavioral measures requires acceptance of the HCP Restricted Data Use Terms (https://www.humanconnectome.org/study/hcp-young-adult/document/restricted-data-usage). The independent validation dataset used to assess the reproducibility of the parcellation homogeneity results is currently not publicly available. The new atlas is openly available in the form of NIFTI (Neuroimaging Informatics Technology Initiative) and CIFTI (Connectivity Informatics Technology Initiative) files. To facilitate mapping of whole-brain connectomes, we have also integrated the new atlas into several well-known cortex-only parcellation atlases and the combined cortex–subcortex atlases are made openly available. Supplementary Table 5 provides details about the atlas formats that can be downloaded from the GitHub repository (https://github.com/yetianmed/subcortex).
Code availability
The Matlab (R2018b) codes to compute Laplacian eigenmaps, gradient magnitudes, diversity curves and other computational analyses undertaken as part of this study are openly available at https://github.com/yetianmed/subcortex. The following additional software packages used for this study are freely and openly available: Diffusion Toolkit (v.0.6.4.1) and TrackVis (v.0.6.1): http://trackvis.org/; NBS (v.1.2): https://www.nitrc.org/projects/nbs/; Icasso (v.1.21): https://research.ics.aalto.fi/ica/icasso/; NeuroMArVL: https://immersive.erc.monash.edu/neuromarvl/; PALM (v.alpha116): https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/PALM/ExchangeabilityBlocks; and fMRIPrep (v.1.5.9): https://fmriprep.org/en/stable/index.html.
References
Eickhoff, S. B., Yeo, B. T. T. & Genon, S. Imaging-based parcellations of the human brain. Nat. Rev. Neurosci. 19, 672–686 (2018).
Glasser, M. F. et al. A multi-modal parcellation of human cerebral cortex. Nature 536, 171–178 (2016).
Yeo, B. T. et al. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J. Neurophysiol. 106, 1125–1165 (2011).
Shepherd, G. M. Corticostriatal connectivity and its role in disease. Nat. Rev. Neurosci. 14, 278–291 (2013).
Choi, E. Y., Yeo, B. T. & Buckner, R. L. The organization of the human striatum estimated by intrinsic functional connectivity. J. Neurophysiol. 108, 2242–2263 (2012).
Ji, J. L. et al. Mapping the human brain’s cortical–subcortical functional network organization. NeuroImage 185, 35–57 (2019).
Cohen, A. L. et al. Defining functional areas in individual human brains using resting functional connectivity MRI. NeuroImage 41, 45–57 (2008).
Gordon, E. M. et al. Generation and evaluation of a cortical area parcellation from resting-state correlations. Cereb. Cortex 26, 288–303 (2016).
Schaefer, A. et al. Local–global parcellation of the human cerebral cortex from intrinsic functional connectivity MRI. Cereb. Cortex 28, 3095–3114 (2018).
Huntenburg, J. M., Bazin, P. L. & Margulies, D. S. Large-scale gradients in human cortical organization. Trends Cogn. Sci. 22, 21–31 (2018).
Haak, K. V., Marquand, A. F. & Beckmann, C. F. Connectopic mapping with resting-state fMRI. NeuroImage 170, 83–94 (2018).
Margulies, D. S. et al. Situating the default-mode network along a principal gradient of macroscale cortical organization. Proc. Natl Acad. Sci. USA 113, 12574–12579 (2016).
Guell, X., Schmahmann, J. D., Gabrieli, J. & Ghosh, S. S. Functional gradients of the cerebellum. eLife 7, e36652 (2018).
King, M., Hernandez-Castillo, C. R., Poldrack, R. A., Ivry, R. B. & Diedrichsen, J. Functional boundaries in the human cerebellum revealed by a multi-domain task battery. Nat. Neurosci. 22, 1371–1378 (2019).
Tian, Y. & Zalesky, A. Characterizing the functional connectivity diversity of the insula cortex: subregions, diversity curves and behavior. NeuroImage 183, 716–733 (2018).
Coalson, T. S., Van Essen, D. C. & Glasser, M. F. The impact of traditional neuroimaging methods on the spatial localization of cortical areas. Proc. Natl Acad. Sci. USA 115, E6356–E6365 (2018).
Fox, M. D. et al. The human brain is intrinsically organized into dynamic, anticorrelated functional networks. Proc. Natl Acad. Sci. USA 102, 9673–9678 (2005).
Van Essen, D. C. et al. The WU–Minn Human Connectome Project: an overview. NeuroImage 80, 62–79 (2013).
Finn, E. S. et al. Functional connectome fingerprinting: identifying individuals using patterns of brain connectivity. Nat. Neurosci. 18, 1664 (2015).
Müller, E. et al. Core and matrix thalamic sub-populations relate to spatio-temporal cortical connectivity gradients. NeuroImage 222, 117224 (2020).
Poppenk, J., Evensmoen, H. R., Moscovitch, M. & Nadel, L. Long-axis specialization of the human hippocampus. Trends Cogn. Sci. 17, 230–240 (2013).
Amunts, K. et al. Cytoarchitectonic mapping of the human amygdala, hippocampal region and entorhinal cortex: intersubject variability and probability maps. Anat. Embryol. 210, 343–352 (2005).
Marquand, A. F., Haak, K. V. & Beckmann, C. F. Functional corticostriatal connection topographies predict goal directed behaviour in humans. Nat. Hum. Behav. 1, 0146 (2017).
Vu, A. et al. Tradeoffs in pushing the spatial resolution of fMRI for the 7T Human Connectome Project. NeuroImage 154, 23–32 (2017).
Gordon, E. M. et al. Precision functional mapping of individual human brains. Neuron 95, 791–807.e7 (2017).
Kong, R. et al. Spatial topography of individual-specific cortical networks predicts human cognition, personality, and emotion. Cereb. Cortex 29, 2533–2551 (2019).
Salehi, M. et al. There is no single functional atlas even for a single individual: functional parcel definitions change with task. NeuroImage 208, 116366 (2020).
Zalesky, A., Fornito, A. & Bullmore, E. T. Network-based statistic: identifying differences in brain networks. NeuroImage 53, 1197–1207 (2010).
Maia, T. V. & Frank, M. J. From reinforcement learning models to psychiatric and neurological disorders. Nat. Neurosci. 14, 154–162 (2011).
Forstmann, B. U., de Hollander, G., van Maanen, L., Alkemade, A. & Keuken, M. C. Towards a mechanistic understanding of the human subcortex. Nat. Rev. Neurosci. 18, 57–65 (2016).
Vos de Wael, R. et al. Anatomical and microstructural determinants of hippocampal subfield functional connectome embedding. Proc. Natl Acad. Sci. USA 115, 10154–10159 (2018).
Churchland, P. S. & Sejnowski, T. J. Perspectives on cognitive neuroscience. Science 242, 741–745 (1988).
Fan, L. et al. The Human Brainnetome Atlas: a new brain atlas based on connectional architecture. Cereb. Cortex 26, 3508–3526 (2016).
Pauli, W. M., Nili, A. N. & Tyszka, J. M. A high-resolution probabilistic in vivo atlas of human subcortical brain nuclei. Sci. Data 5, 180063 (2018).
Plachti, A. et al. Multimodal parcellations and extensive behavioral profiling tackling the hippocampus gradient. Cereb. Cortex 29, 4595–4612 (2019).
Janssen, R. J., Jylänki, P., Kessels, R. P. C. & van Gerven, M. A. J. Probabilistic model-based functional parcellation reveals a robust, fine-grained subdivision of the striatum. NeuroImage 119, 398–405 (2015).
Behrens, T. E. et al. Non-invasive mapping of connections between human thalamus and cortex using diffusion imaging. Nat. Neurosci. 6, 750–757 (2003).
Haber, S. N. & Knutson, B. The reward circuit: linking primate anatomy and human imaging. Neuropsychopharmacology 35, 4–26 (2010).
Morel, A., Magnin, M. & Jeanmonod, D. Multiarchitectonic and stereotactic atlas of the human thalamus. J. Comp. Neurol. 387, 588–630 (1997).
Meredith, G. E., Pattiselanno, A., Groenewegen, H. J. & Haber, S. N. Shell and core in monkey and human nucleus accumbens identified with antibodies to calbindin-D28k. J. Comp. Neurol. 365, 628–639 (1996).
Cocchi, L. & Zalesky, A. Personalized transcranial magnetic stimulation in psychiatry. Biol. Psychiatry Cogn. Neurosci. Neuroimaging 3, 731–741 (2018).
Castro, D. C. & Bruchas, M. R. A motivational and neuropeptidergic hub: anatomical and functional diversity within the nucleus accumbens shell. Neuron 102, 529–552 (2019).
Pauli, W. M., O’Reilly, R. C., Yarkoni, T. & Wager, T. D. Regional specialization within the human striatum for diverse psychological functions. Proc. Natl Acad. Sci. USA 113, 1907–1912 (2016).
MacAskill, A. F., Little, J. P., Cassel, J. M. & Carter, A. G. Subcellular connectivity underlies pathway-specific signaling in the nucleus accumbens. Nat. Neurosci. 15, 1624–1626 (2012).
Perez, S. M. & Lodge, D. J. Convergent inputs from the hippocampus and thalamus to the nucleus accumbens regulate dopamine neuron activity. J. Neurosci. 38, 10607–10618 (2018).
Pontieri, F. E., Tanda, G., Orzi, F. & Di Chiara, G. Effects of nicotine on the nucleus accumbens and similarity to those of addictive drugs. Nature 382, 255–257 (1996).
Gallinat, J. et al. Abnormal hippocampal neurochemistry in smokers: evidence from proton magnetic resonance spectroscopy at 3T. J. Clin. Psychopharmacol. 27, 80–84 (2007).
Xu, Z., Seidler, F. J., Ali, S. F., Slikker, W. Jr. & Slotkin, T. A. Fetal and adolescent nicotine administration: effects on CNS serotonergic systems. Brain Res. 914, 166–178 (2001).
de Leon, J. & Diaz, F. J. A meta-analysis of worldwide studies demonstrates an association between schizophrenia and tobacco smoking behaviors. Schizophrenia Res. 76, 135–157 (2005).
Koukouli, F. et al. Nicotine reverses hypofrontality in animal models of addiction and schizophrenia. Nat. Med. 23, 347–354 (2017).
Winkler, A. M., Ridgway, G. R., Webster, M. A., Smith, S. M. & Nichols, T. E. Permutation inference for the general linear model. NeuroImage 92, 381–397 (2014).
Winkler, A. M., Webster, M. A., Vidaurre, D., Nichols, T. E. & Smith, S. M. Multi-level block permutation. NeuroImage 123, 253–268 (2015).
Smith, S. M. et al. Resting-state fMRI in the Human Connectome Project. NeuroImage 80, 144–168 (2013).
Barch, D. M. et al. Function in the human connectome: task-fMRI and individual differences in behavior. NeuroImage 80, 169–189 (2013).
Glasser, M. F. et al. The minimal preprocessing pipelines for the Human Connectome Project. NeuroImage 80, 105–124 (2013).
Salimi-Khorshidi, G. et al. Automatic denoising of functional MRI data: combining independent component analysis and hierarchical fusion of classifiers. NeuroImage 90, 449–468 (2014).
Griffanti, L. et al. ICA-based artefact removal and accelerated fMRI acquisition for improved resting state network imaging. NeuroImage 95, 232–247 (2014).
Parkes, L., Fulcher, B., Yucel, M. & Fornito, A. An evaluation of the efficacy, reliability, and sensitivity of motion correction strategies for resting-state functional MRI. NeuroImage 171, 415–436 (2018).
Power, J. D., Barnes, K. A., Snyder, A. Z., Schlaggar, B. L. & Petersen, S. E. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. NeuroImage 59, 2142–2154 (2012).
Satterthwaite, T. D. et al. An improved framework for confound regression and filtering for control of motion artifact in the preprocessing of resting-state functional connectivity data. NeuroImage 64, 240–256 (2013).
Van Dijk, K. R., Sabuncu, M. R. & Buckner, R. L. The influence of head motion on intrinsic functional connectivity MRI. NeuroImage 59, 431–438 (2012).
Puckett, A. M. et al. Using multi-echo simultaneous multi-slice (SMS) EPI to improve functional MRI of the subcortical nuclei of the basal ganglia at ultra-high field (7T). NeuroImage 172, 886–895 (2018).
Glasser, M. F. et al. The Human Connectome Project’s neuroimaging approach. Nat. Neurosci. 19, 1175–1187 (2016).
Glasser, M. F. et al. Using temporal ICA to selectively remove global noise while preserving global signal in functional MRI data. NeuroImage 181, 692–717 (2018).
Cole, M. W. et al. Multi-task connectivity reveals flexible hubs for adaptive task control. Nat. Neurosci. 16, 1348–1355 (2013).
Esteban, O. et al. fMRIPrep: a robust preprocessing pipeline for functional MRI. Nat. Methods 16, 111–116 (2019).
Percheron, G., Yelnik, J. & Francois, C. A Golgi analysis of the primate globus pallidus. III. Spatial organization of the striato–pallidal complex. J. Comp. Neurol. 227, 214–227 (1984).
Yushkevich, P. A. et al. Quantitative comparison of 21 protocols for labeling hippocampal subfields and parahippocampal subregions in in vivo MRI: towards a harmonized segmentation protocol. NeuroImage 111, 526–541 (2015).
Guadalupe, T. et al. Human subcortical brain asymmetries in 15,847 people worldwide reveal effects of age and sex. Brain Imaging Behav. 11, 1497–1514 (2017).
de Hollander, G., Keuken, M. C., van der Zwaag, W., Forstmann, B. U. & Trampel, R. Comparing functional MRI protocols for small, iron-rich basal ganglia nuclei such as the subthalamic nucleus at 7 T and 3 T. Hum. Brain Mapp. 38, 3226–3248 (2017).
Belkin, M. & Niyogi, P. Laplacian eigenmaps for dimensionality reduction and data representation. Neural Comput. 15, 1373–1396 (2003).
Fornito, A., Zalesky, A. & Breakspear, M. Graph analysis of the human connectome: promise, progress, and pitfalls. NeuroImage 80, 426–444 (2013).
Cerliani, L. et al. Probabilistic tractography recovers a rostrocaudal trajectory of connectivity variability in the human insular cortex. Hum. Brain Mapp. 33, 2005–2034 (2012).
Jeurissen, B., Descoteaux, M., Mori, S. & Leemans, A. Diffusion MRI fiber tractography of the brain. NMR Biomed. 32, e3785 (2019).
Alexander, D. C. in Visualization and Processing of Tensor Fields (eds Weickert J. & Hagen, H.) Ch. 5 (Springer, 2006).
Friman, O., Farneback, G. & Westin, C. F. A Bayesian approach for stochastic white matter tractography. IEEE Trans. Med. Imaging 25, 965–978 (2006).
Peled, S., Friman, O., Jolesz, F. & Westin, C. F. Geometrically constrained two-tensor model for crossing tracts in DWI. Magn. Reson. Imaging 24, 1263–1270 (2006).
Behrens, T. E., Berg, H. J., Jbabdi, S., Rushworth, M. F. & Woolrich, M. W. Probabilistic diffusion tractography with multiple fibre orientations: what can we gain? NeuroImage 34, 144–155 (2007).
Wang, R., Benner, T., Sorensen, A. & Wedeen, V. Diffusion Toolkit: a software package for diffusion imaging data processing and tractography. Proc. Int. Soc. Magn. Reson. Med. 15, 3720 (2007).
O’Donnell, L. J. & Westin, C. F. Automatic tractography segmentation using a high-dimensional white matter atlas. IEEE Trans. Med. Imaging 26, 1562–1575 (2007).
Müller, M. in Information Retrieval for Music and Motion 69–84 (Springer, 2007).
Benjamini, Y. Discovering the false discovery rate. J. R. Stat. Soc. Ser. B 72, 405–416 (2010).
Serrano, M. Á., Boguñá, M. & Vespignani, A. Extracting the multiscale backbone of complex weighted networks. Proc. Natl Acad. Sci. USA 106, 6483–6488 (2009).
Meyer, F. Topographic distance and watershed lines. Signal Process. 38, 113–125 (1994).
Eickhoff, S. B. et al. A new SPM toolbox for combining probabilistic cytoarchitectonic maps and functional imaging data. NeuroImage 25, 1325–1335 (2005).
Lancichinetti, A., Fortunato, S. & Kertész, J. Detecting the overlapping and hierarchical community structure in complex networks. N. J. Phys. 11, 033015 (2009).
Greene, A. S., Gao, S., Scheinost, D. & Constable, R. T. Task-induced brain state manipulation improves prediction of individual traits. Nat. Commun. 9, 2807 (2018).
Cole, M. W., Bassett, D. S., Power, J. D., Braver, T. S. & Petersen, S. E. Intrinsic and task-evoked network architectures of the human brain. Neuron 83, 238–251 (2014).
Medaglia, J. D., Lynall, M. E. & Bassett, D. S. Cognitive network neuroscience. J. Cogn. Neurosci. 27, 1471–1491 (2015).
Smith, S. M. et al. A positive-negative mode of population covariation links brain connectivity, demographics and behavior. Nat. Neurosci. 18, 1565–1567 (2015).
Fornito, A., Zalesky, A., Pantelis, C. & Bullmore, E. T. Schizophrenia, neuroimaging and connectomics. NeuroImage 62, 2296–2314 (2012).
Xia, C. H. et al. Linked dimensions of psychopathology and connectivity in functional brain networks. Nat. Commun. 9, 3003 (2018).
Blondel, V. D., Guillaume, J.-L., Lambiotte, R. & Lefebvre, E. Fast unfolding of communities in large networks. J. Stat. Mech. Theory Exp. 2008, P10008 (2008).
Van der Waerden, B. Oder tests for two-sample problem and their power. Proc. K. Nederlandse Akademie van Wetenschappen. Ser. A 55, 453–458 (1952).
Himberg, J., Hyvarinen, A. & Esposito, F. Validating the independent components of neuroimaging time series via clustering and visualization. NeuroImage 22, 1214–1222 (2004).
Hyvarinen, A. Fast and robust fixed-point algorithms for independent component analysis. IEEE Trans. Neural Netw. 10, 626–634 (1999).
Kuhn, H. W. The Hungarian method for the assignment problem. Nav. Res. Logist. Q 2, 83–97 (1955).
Freedman, D. & Lane, D. A nonstochastic interpretation of reported significance levels. J. Bus. Economic Stat. 1, 292–298 (1983).
Phillips, J. W. et al. A repeated molecular architecture across thalamic pathways. Nat. Neurosci. 22, 1925–1935 (2019).
Hwang, K., Bertolero, M. A., Liu, W. B. & D’Esposito, M. The human thalamus is an integrative hub for functional brain networks. J. Neurosci. 37, 5594–5607 (2017).
Shine, J. M. et al. The low-dimensional neural architecture of cognitive complexity is related to activity in medial thalamic nuclei. Neuron 104, 849–855.e3 (2019).
Mink, J. W. The basal ganglia: focused selection and inhibition of competing motor programs. Prog. Neurobiol. 50, 381–425 (1996).
Janak, P. H. & Tye, K. M. From circuits to behaviour in the amygdala. Nature 517, 284–292 (2015).
Acknowledgements
We thank M. Glasser (Washington University) for provision of the Wishart filter code. We thank L. Cocchi (QIMR Berghofer Medical Research Institute) for provision of the additional validation dataset. All other data were provided by the HCP, the WU–Minn Consortium (1U54MH091657; Principal Investigators: D. Van Essen and K. Ugurbil) funded by the 16 National Institutes of Health (NIH) institutes and centers that support the NIH Blueprint for Neuroscience Research, and by the McDonnell Center for Systems Neuroscience at Washington University. Y.T. was supported by a NHMRC Project Grant awarded to A.Z. (APP1142801). M.B. and A.Z. were each supported by research fellowships from the NHMRC (APP1136649 and APP1118153, respectively).
Author information
Authors and Affiliations
Contributions
Y.T. and A.Z. conceived the idea and designed the study. Y.T. compiled the data, performed the analyses and prepared the visualizations. Y.T. and A.Z. drafted the manuscript. D.S.M. and M.B. provided critical conceptual input. All authors provided critical feedback and editing of the final manuscript. A.Z. supervised the research.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Neuroscience thanks Janine Bijsterbosch, Rodrigo Braga and Evan Gordon for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–24 and Supplementary Tables 1–5.
Rights and permissions
About this article
Cite this article
Tian, Y., Margulies, D.S., Breakspear, M. et al. Topographic organization of the human subcortex unveiled with functional connectivity gradients. Nat Neurosci 23, 1421–1432 (2020). https://doi.org/10.1038/s41593-020-00711-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41593-020-00711-6
This article is cited by
-
Dynamic functional changes upon thalamotomy in essential tremor depend on baseline brain morphometry
Scientific Reports (2024)
-
Nuclei-specific hypothalamus networks predict a dimensional marker of stress in humans
Nature Communications (2024)
-
High-intensity sweet taste as a predictor of subjective alcohol responses to the ascending limb of an intravenous alcohol prime: an fMRI study
Neuropsychopharmacology (2024)
-
Blunted brain responses to neutral faces in healthy first-degree relatives of patients with schizophrenia: an image-based fMRI meta-analysis
Schizophrenia (2024)
-
Whole-brain dynamics across the menstrual cycle: the role of hormonal fluctuations and age in healthy women
npj Women's Health (2024)