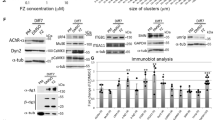
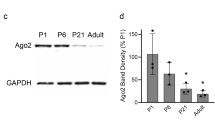
Abstract
Neuromuscular junction (NMJ) disruption is an early pathogenic event in amyotrophic lateral sclerosis (ALS). Yet, direct links between NMJ pathways and ALS-associated genes such as FUS, whose heterozygous mutations cause aggressive forms of ALS, remain elusive. In a knock-in Fus-ALS mouse model, we identified postsynaptic NMJ defects in newborn homozygous mutants that were attributable to mutant FUS toxicity in skeletal muscle. Adult heterozygous knock-in mice displayed smaller neuromuscular endplates that denervated before motor neuron loss, which is consistent with ‘dying-back’ neuronopathy. FUS was enriched in subsynaptic myonuclei, and this innervation-dependent enrichment was distorted in FUS-ALS. Mechanistically, FUS collaborates with the ETS transcription factor ERM to stimulate transcription of acetylcholine receptor genes. Co-cultures of induced pluripotent stem cell-derived motor neurons and myotubes from patients with FUS-ALS revealed endplate maturation defects due to intrinsic FUS toxicity in both motor neurons and myotubes. Thus, FUS regulates acetylcholine receptor gene expression in subsynaptic myonuclei, and muscle-intrinsic toxicity of ALS mutant FUS may contribute to dying-back motor neuronopathy.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The data that support the findings of this study and biological materials used are available from the corresponding authors upon request.
References
Darabid, H., Perez-Gonzalez, A. P. & Robitaille, R. Neuromuscular synaptogenesis: coordinating partners with multiple functions. Nat. Rev. Neurosci. 15, 703–718 (2014).
Tintignac, L. A., Brenner, H. R. & Ruegg, M. A. Mechanisms regulating neuromuscular junction development and function and causes of muscle wasting. Physiol. Rev. 95, 809–852 (2015).
Shi, L., Fu, A. K. & Ip, N. Y. Molecular mechanisms underlying maturation and maintenance of the vertebrate neuromuscular junction. Trends Neurosci. 35, 441–453 (2012).
Hippenmeyer, S., Huber, R. M., Ladle, D. R., Murphy, K. & Arber, S. ETS transcription factor Erm controls subsynaptic gene expression in skeletal muscles. Neuron 55, 726–740 (2007).
Ravel-Chapuis, A., Vandromme, M., Thomas, J. L. & Schaeffer, L. Postsynaptic chromatin is under neural control at the neuromuscular junction. EMBO J. 26, 1117–1128 (2007).
Taylor, J. P., Brown, R. H. Jr & Cleveland, D. W. Decoding ALS: from genes to mechanism. Nature 539, 197–206 (2016).
Pun, S., Santos, A. F., Saxena, S., Xu, L. & Caroni, P. Selective vulnerability and pruning of phasic motoneuron axons in motoneuron disease alleviated by CNTF. Nat. Neurosci. 9, 408–419 (2006).
Fischer, L. R. et al. Amyotrophic lateral sclerosis is a distal axonopathy: evidence in mice and man. Exp. Neurol. 185, 232–240 (2004).
Dadon-Nachum, M., Melamed, E. & Offen, D. The ‘dying-back’ phenomenon of motor neurons in ALS. J. Mol. Neurosci. 43, 470–477 (2011).
Schwartz, J. C., Cech, T. R. & Parker, R. R. Biochemical properties and biological functions of FET proteins. Annu. Rev. Biochem. 84, 355–379 (2015).
Ling, S. C., Polymenidou, M. & Cleveland, D. W. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron 79, 416–438 (2013).
Deng, H., Gao, K. & Jankovic, J. The role of FUS gene variants in neurodegenerative diseases. Nat. Rev. Neurol. 10, 337–348 (2014).
Dormann, D. et al. ALS-associated fused in sarcoma (FUS) mutations disrupt transportin-mediated nuclear import. EMBO J. 29, 2841–2857 (2010).
Scekic-Zahirovic, J. et al. Toxic gain of function from mutant FUS protein is crucial to trigger cell autonomous motor neuron loss. EMBO J. 35, 1077–1097 (2016).
Scekic-Zahirovic, J. et al. Motor neuron intrinsic and extrinsic mechanisms contribute to the pathogenesis of FUS-associated amyotrophic lateral sclerosis. Acta Neuropathol. 133, 887–906 (2017).
Kanisicak, O., Mendez, J. J., Yamamoto, S., Yamamoto, M. & Goldhamer, D. J. Progenitors of skeletal muscle satellite cells express the muscle determination gene, MyoD. Dev. Biol. 332, 131–141 (2009).
Yamamoto, M. et al. A multifunctional reporter mouse line for Cre- and FLP-dependent lineage analysis. Genesis 47, 107–114 (2009).
Muzumdar, M. D., Tasic, B., Miyamichi, K., Li, L. & Luo, L. A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605 (2007).
Tan, A. Y., Riley, T. R., Coady, T., Bussemaker, H. J. & Manley, J. L. TLS/FUS (translocated in liposarcoma/fused in sarcoma) regulates target gene transcription via single-stranded DNA response elements. Proc. Natl Acad. Sci. USA 109, 6030–6035 (2012).
Higelin, J. et al. FUS mislocalization and vulnerability to DNA damage in ALS patients derived hiPSCs and aging motoneurons. Front. Cell Neurosci. 10, 290 (2016).
Hosoyama, T., McGivern, J. V., Van Dyke, J. M., Ebert, A. D. & Suzuki, M. Derivation of myogenic progenitors directly from human pluripotent stem cells using a sphere-based culture. Stem Cells Transl Med. 3, 564–574 (2014).
Demestre, M. et al. Formation and characterisation of neuromuscular junctions between hiPSC derived motoneurons and myotubes. Stem Cell Res. 15, 328–336 (2015).
de Carvalho, M. et al. Electrodiagnostic criteria for diagnosis of ALS. Clin. Neurophysiol. 119, 497–503 (2008).
So, E. et al. Mitochondrial abnormalities and disruption of the neuromuscular junction precede the clinical phenotype and motor neuron loss in hFUSWT transgenic mice. Hum. Mol. Genet. 27, 463–474 (2018).
Sharma, A. et al. ALS-associated mutant FUS induces selective motor neuron degeneration through toxic gain of function. Nat. Commun. 7, 10465 (2016).
Naumann, M. et al. Impaired DNA damage response signaling by FUS-NLS mutations leads to neurodegeneration and FUS aggregate formation. Nat. Commun. 9, 335 (2018).
Miller, T. M. et al. Gene transfer demonstrates that muscle is not a primary target for non-cell-autonomous toxicity in familial amyotrophic lateral sclerosis. Proc. Natl Acad. Sci. USA 103, 19546–19551 (2006).
Towne, C., Raoul, C., Schneider, B. L. & Aebischer, P. Systemic AAV6 delivery mediating RNA interference against SOD1: neuromuscular transduction does not alter disease progression in fALS mice. Mol. Ther. 16, 1018–1025 (2008).
Dobrowolny, G. et al. Skeletal muscle is a primary target of SOD1G93A-mediated toxicity. Cell Metab. 8, 425–436 (2008).
Wong, M. & Martin, L. J. Skeletal muscle-restricted expression of human SOD1 causes motor neuron degeneration in transgenic mice. Hum. Mol. Genet. 19, 2284–2302 (2010).
Williams, A. H. et al. MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science 326, 1549–1554 (2009).
Kedage, V. et al. An Interaction with Ewing’s sarcoma breakpoint protein EWS defines a specific oncogenic mechanism of ETS factors rearranged in prostate cancer. Cell Rep. 17, 1289–1301 (2016).
Vandesompele, J. et al. Accurate normalization of real-time quantitative RT–PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, RESEARCH0034 (2002).
Belzil, V. V. et al. Novel FUS deletion in a patient with juvenile amyotrophic lateral sclerosis. Arch. Neurol. 69, 653–656 (2012).
Japtok, J. et al. Stepwise acquirement of hallmark neuropathology in FUS-ALS iPSC models depends on mutation type and neuronal aging. Neurobiol. Dis. 82, 420–429 (2015).
Lenzi, J. et al. ALS mutant FUS proteins are recruited into stress granules in induced pluripotent stem cell-derived motoneurons. Dis. Model Mech. 8, 755–766 (2015).
Aasen, T. et al. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat. Biotechnol. 26, 1276–1284 (2008).
Takahashi, K. & Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006).
Warlich, E. et al. Lentiviral vector design and imaging approaches to visualize the early stages of cellular reprogramming. Mol. Ther. 19, 782–789 (2011).
Stockmann, M. et al. Developmental and functional nature of human iPSC derived motoneurons. Stem Cell Rev. 9, 475–492 (2013).
Linta, L. et al. Rat embryonic fibroblasts improve reprogramming of human keratinocytes into induced pluripotent stem cells. Stem Cells Dev. 21, 965–976 (2012).
Hu, B. Y. & Zhang, S. C. Differentiation of spinal motor neurons from pluripotent human stem cells. Nat. Protoc. 4, 1295–1304 (2009).
Darabi, R. et al. Human ES- and iPS-derived myogenic progenitors restore DYSTROPHIN and improve contractility upon transplantation in dystrophic mice. Cell Stem Cell 10, 610–619 (2012).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Bischoff, C., Stalberg, E., Falck, B. & Eeg-Olofsson, K. E. Reference values of motor unit action potentials obtained with multi-MUAP analysis. Muscle Nerve 17, 842–851 (1994).
Gilai, A. N. in Computer-aided Electromyography and Expert Systems, Clinical Neurophysiology Updates (ed. Desmedt, J. E.) 143–161 (Karger, 1989).
Acknowledgements
The authors thank L. Schaeffer (University of Lyon, France), A. Verger (Université Lille Nord de France, France), J. Weishaupt (Ulm University, Germany) and R. Perlingeiro (University of Minnesota, USA) for the gift of plasmids, and N. Charlet Berguerand for advice on siRNA treatment. They would also like to thank the General Instruments Facility of the Faculty of Science of Radboud University and I. Alexopoulos in particular for advice on image acquisition and analyses. The authors would like to thank S. Seltenheim, L. Dietz and R. Zienecker for excellent technical support, A. Knehr (Department of Neurology, Ulm University) for collecting human material, and B. Mayer (Institute for Epidemiology and Medical Biometrics, Ulm University) for advice on statistical analyses. They are grateful to all the participants for willingly providing them with hair and muscle samples. This work was directly supported by the following grants: ALS Association Investigator Initiated Awards (grants 2235, 3209 and 8075 to L.D. and C.L.-T.); the Frick Foundation (award 2013 to L.D. and C.L.-T); Association Française contre les Myopathies (grant 18280 to L.D., C.L.-T. and E.S.); Virtual Helmholtz Institute ‘RNA dysmetabolism in ALS and FTD’ (WP2 to L.D., A.L. and T.M.B.); the DZNE (Ulm site) and agence nationale de la recherche (ToFU, EpiFUS to L.D.); the Bundesministerium für Bildung und Forschung (BMBF01EK1611C to T.M.B. and M.D.); the Max Planck Society (to E.S.); and the Donders Center for Neuroscience (to E.S.). Work in the authors’ laboratories is supported by ARSla (call 2014 and 2016 to L.D.), the fondation ‘recherche sur le cerveau’ (call 2015 to L.D.), Axa Banque Patrimoniale (Bourse recherche maladies rares to L.D.), Fondation pour la recherche médicale (Equipe FRM to L.D.), the Muscular Dystrophy Association (MDA479773 to E.S.), the EU Joint Programme—Neurodegenerative Disease Research (JPND; grant numbers ZonMW 733051075 (TransNeuro) and ZonMW 733501073 (LocalNMD) to E.S.) and an ERC consolidator grant (ERC-2017-COG 770244 to E.S).
Author information
Authors and Affiliations
Contributions
G.P. performed most of the experiments in heterozygous knock-in mice and in C2C12 cells, with the help of S.D., M.-A.G., J.S.-Z. and I.S.-R. M.D., J.H. and A.A. performed most of the experiments in human-derived cells and tissues. A.Z., M.B., M.M., L.Z., C. Sijlmans, S.M., M.W., N.v.B. and E.S. performed and analyzed the experiments in homozygous knock-in, MyoD-Cre rescued knock-in and knock-out mice. C. Sellier performed immunoprecipitation experiments. N.M. performed electron microscopy. A.L.-B supervised the ChIP experiments. A.R. provided human muscle biopsy samples. A.H. performed and analyzed the patient EMG analyses. A.L. provided patient and clinical material. M.D., C.L.-T., T.M.B., L.D. and E.S. initiated, conceived and supervised the project. G.P., M.D., L.D. and E.S. wrote the manuscript. All authors contributed to the experimental design and interpretation and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Neuroscience thanks Thomas Lloyd and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 NMJ morphology defects in the gastrocnemius of newborn FusΔNLS/ΔNLS and Fus−/− mice.
a-f Dot plots showing the percentage (%) of innervated endplates (a, d), endplate surface area as % of Fus+/+ littermate controls (b, e) and the total number of endplates in the gastrocnemius (c, f) of newborn FusΔNLS/ΔNLS (a-c, closed circles) and Fus−/− (d-f, closed circles) mice, as compared to Fus+/+ mice (open circles). Two-tailed unpaired t-test; **p<0.01, ***p=0.0005; N= 12 versus 11 (a), 17 versus 14 (b), 4 versus 5 (c), 8 versus 8 (d), 15 versus 13 (e), 9 versus 10 (f) mice. Average ± SEM. g-k, Quantification of ultrastructural NMJ defects in gastrocnemius of E18.5 Fus+/+, Fus−/− and FusΔNLS/ΔNLS mice. g, The neuromuscular contact was scored as intact (1) when pre- and postsynaptic membranes were closely apposed and the synaptic cleft was filled with electron dense material. A widened synaptic cleft was scored as 0, and intermediate morphology as 0.5. h, The presence or absence of postsynaptic membrane foldings was scored as 1 or 0, respectively. i,j, Presynaptic (i) and postsynaptic (j) membrane integrity was scored as intact (1), thin (0.5) or disrupted (0). k, The abundance of synaptic vesicles was scored as abundant (1), rare (0.5) or absent (0). For g-k: Kruskal-Wallis test with Dunn’s multiple comparisons test; *p<0.05, **p<0.01; N= 5 Fus+/+, 4 Fus−/− and 5 FusΔNLS/ΔNLS mice; for each parameter, the average value per animal (based on 4-14 NMJs analyzed per animal) was used as a data point. Average ± SEM.
Supplementary Figure 2 Quantification of myofiber number in hind limb muscles of Fus mutant mice.
a,b, Dystrophin staining to visualize individual muscle fibers in extensor digitorum longus (EDL) muscle of newborn Fus+/+ versus FusΔNLS/ΔNLS mice. Scale bar: 100µm. c,d, Dot plots showing the total number of muscle fibers in EDL muscle of newborn Fus+/+ (open circles) versus FusΔNLS/ΔNLS (c, closed circles) or Fus−/− (d, closed circles) mice. p=0.52 (c) by two-tailed unpaired t-test; N= 6 versus 5 (c) and 2 versus 2 (d) mice. Average ± SEM. e, Dot plot showing the total number of muscle fibers in TA of newborn Fus+/+ (open circles) versus Fus−/− (closed circles) mice. p=0.054 by two-tailed unpaired t-test; N= 7 versus 4 mice. Average ± SEM. f, Dot plot showing the total number of endplates in TA of 1-month-old male Fus+/+ versus FusΔNLS/+ mice. p=0.59 by two-tailed unpaired t-test; N= 3 versus 7 mice. Average ± SEM.
Supplementary Figure 3 MyoDiCre mediates highly efficient recombination between LoxP sites.
a, Schematic illustrating the working principle of the ROSAmT/mG reporter mice. The chicken β-actin core promoter with a CMV enhancer (pCA) drives expression of the loxP-flanked (triangles) coding sequence of membrane-targeted tandem dimer Tomato (mT). After Cre-mediated excision of the mT sequence, the pCA promoter drives expression of membrane-targeted EGFP (mG). pA: polyadenylation sequence. b, Transverse sections through the TA of adult ROSAmT/mG/+ (control, upper panels) and MyoDiCre/+; ROSAmT/mG/+ (experimental, lower panels) mice were labeled with phalloidin (to visualize muscle fibers), antibodies against tdTomato and GFP, and DAPI. Scale bar: 20µm. c, Bar graph showing the percentage (%) or GFP-positive muscle fibers in ROSAmT/mG/+ and MyoDiCre/+; ROSAmT/mG/+ mice. One-sample Wilcoxon signed rank test (mT/mG; MyoD-iCre versus 0); *p=0.031; N= 6 versus 6 mice. Average ± SEM.
Supplementary Figure 4 Reduced expression of Chrn genes in Fus-ALS mouse models.
a,b, Relative mRNA levels of Chrn genes, including Chrna1, Chrnb1, Chrng, Chrnd, and Chrne, in gastrocnemius muscle of E18.5 Fus+/+ versus FusΔNLS/ΔNLS mice (a) and TA muscle of 1-month-old male Fus+/+ versus FusΔNLS/+ mice (b). Note that Chrne is not expressed at E18.5, and Chrng is not expressed at 1 month of age. Two-tailed unpaired t-test; *p<0.05, **p=0.0055; N= 4 Fus+/+ versus 5 FusΔNLS/ΔNLS (a) and 4 Fus+/+ versus 6 FusΔNLS/+ mice (b). Average ± SEM.
Supplementary Figure 5 R495X mutant FUS mislocalizes to the cytoplasm and induces cytoplasmic mislocalization and granular nuclear staining of wild type FUS in C2C12 cells.
a, Representative images of C2C12 cells co-transfected with plasmids expressing N-terminal GFP-tagged wild type human FUS (FUS_WT::GFP) and N-terminal Myc-tagged human FUS, either wild type (FUS_WT::Myc) or R495X mutant (FUS_R495X::Myc). 24 h later, cells were immunostained for Myc and Hoechst was used to label nuclei. Scale bar: 25 µm. b,c, Quantification of the percentage of cytoplasmic relative to total (nuclear + cytoplasmic) FUS_WT::GFP (b) and the percentage of granular relative to total nuclear FUS_WT::GFP (c) in C2C12 cells co-transfected with either FUS_WT::Myc (open circles) or FUS_R495X::Myc (closed circles). Nested one sample Wilcoxon-test (b); ***p=0.0005; Nested t-test (c); *p=0.024; N= 3 transfections, with for each 11-12 images analyzed and 10-50 transfected cells per image. Average ± SEM.
Supplementary Figure 6 FUS and ERM collaborate to promote transcription of Chrn genes in subsynaptic nuclei.
a, FUS is required for Agrin induced transcriptional activation of the Chrne promoter. Luminescence intensity (in RLU) measured in C2C12 cells 24 h after transfection with siCt (25 nM) or siFus (25 nM) and a luciferase reporter plasmid carrying the promoter of Chrne gene, and further treated for 24 h with agrin (10 ng/mL) or its vehicle. Data of 10 independent transfections are shown. Each transfection included 3 to 6 technical replicates per condition. For each transfection, luminescence intensity was normalized to siCt without agrin treatment after removal of statistical outliers, and the median value of the technical replicates was determined per condition, plotted in the graph and used for statistical analysis. Lines and error bars indicate median ± interquartile range. Two-way ANOVA with Tukey post-hoc test for multiple comparisons; **p=0.0041; ***p<0.001; ns: not significant. b, HEK293 cells were transfected with FLAG-ERM and Myc-Fus expression plasmids and lysates were immunoblotted before (input) and after (IP) immunoprecipitation with anti-Myc, in the presence or absence of RNase. c, Endogenous FUS was immunoprecipitated from HEK293 cells using a specific antibody (IP) or beads only, in the presence or absence of RNase. Native lysates (input) and immunoprecipitates were analyzed by Western blotting with anti-FUS or anti-ERM antibodies. Co-IP experiments (b, c) were independently repeated 5 (b) or 6 (c) times with similar results. Blot images in panels b and c were cropped. Supplementary Fig. 10 shows uncropped blots.
Supplementary Figure 7 Characterization of hiPSC-derived myotubes.
a, Time course of the spheres-based protocol for myogenic differentiation, including progenitor and final myotube differentiation steps. Myogenic cells were grown in spheres for 6 weeks and then plated to form multinucleated myotubes. b, Relative mRNA levels of the myogenic markers MYOG, PAX7 and CHRNA1 as determined by quantitative real time PCR (qPCR) on hiPSC-derived myocytes from two control and two FUS-ALS patients at different time points during differentiation (4, 5, 6, 7 and 8 weeks). The HMBS gene (encoding hydroxymethylbilane synthase) was used as housekeeping gene for normalization. Transcript levels of CHRNA1 were significantly reduced in FUS-ALS myotubes after 8 weeks of differentiation. ***p<0.001 by two-way ANOVA with Bonferroni post-test. N= 3 independent experiments per genotype, with each experiment representing the average of two technical replicates. Average ± SEM. c, Differentiated muscle cells stained positive for the muscle-specific intermediate filament desmin, the contractile filament myosin heavy chain (MHC), MYOG, the skeletal muscle ryanodine receptor isoform RyR1 and the voltage-dependent dihydropyridine receptor alpha subunit (DHPRα). The latter two are intracellular calcium channels that mediate calcium release from the sarcoplasmic reticulum, an essential step in muscle contraction. Staining for FUS and DAPI revealed FUS localization in nuclei. Differentiation was repeated three times with similar results. Scale bar: 10µm. d, hiPSC-derived myotubes were labeled with BTX (AChR, yellow), FUS (green), actinin (red) and DAPI (blue). FUS predominantly localized to nuclei in both CNTL and FUS-ALS myotubes. The microfilament protein α-actinin, which is necessary for attachment of actin filaments to the sarcomeric Z-disks, was correctly localized to Z-disks. Differentiation was repeated three times with similar results. Scale bar: 10 µm.
Supplementary Figure 8 Characterization hiPSC-derived motor neuron-myotube co-cultures.
a,b, Size of single BTX positive puncta within endplates in motor neuron-myotube co-cultures. Compared to CNTL-CNTL co-cultures, combinations containing FUS-ALS myotubes showed reduced single particle size. Panel a shows co-cultures derived from CNTL1 and FUS1 or FUS2, panel b shows co-cultures derived from FUS1 and its isogenic control CNTL3. **p<0.01; ***p<0.001 by Kruskal-Wallis test with Dunn’s test for multiple comparisons. N= 759 (CNTL1 MT + CNTL1 MN), 846 (CNTL1 MT + FUS1 MN), 3467 (FUS1 MT + CNTL1 MN), 1872 (FUS1 MT + FUS1 MN), 307 (CNTL1 MT + FUS2 MN), 364 (FUS2 MT + CNTL1 MN), 841 (FUS2 MT + FUS2 MN), 978 (CNTL3 MT + CNTL3 MN), 1842 (CNTL3 MT + FUS1MN), 1799 (FUS1 MT + CNTL3 MN), 1116 (FUS1 MT + FUS1 MN). N= 3 co-cultures per combination. Average ± SEM. c, Motor neuron-myotube co-cultures were labeled for FUS (green), AChR (BTX, yellow), neurofilament heavy chain (NF-H, red, labels axons) and DAPI (blue). FUS localized to nuclei of both CNTL and FUS myotubes (white arrowheads), but in FUS-mutant motor neurons, FUS localized to both nucleus and cytoplasm, in contrast to nuclear FUS localization in CNTL motor neurons (red arrowheads). Scale bar: 10µm. d, Relative mRNA levels of myogenic differentiation markers (PAX7, MYOD, MYOG) and AChR subunits (CHRNA1, CHRNG, CHRNE) in motor neuron-myotube co-cultures as determined by qPCR. As myotube differentiation was induced by PAX7 overexpression, it was expected that no significant differences in PAX7 expression were found across the different genotypic combinations. The HMBS gene was used as housekeeping gene for normalization. *p<0.05, **p<0.01, ***p<0.001 by one-way ANOVA with Bonferroni post-test. N= 3 independent co-cultures per genotype, with each representing the average of two technical replicates. Average ± SEM.
Supplementary Figure 9 Characterization of muscle biopsies from FUS-ALS patients and controls.
a, Distribution of muscle fiber size in muscle biopsies from control (gray bars) and FUS-ALS patients (green bars). Muscle fibers were assorted in the indicated size categories based on their surface area (in μm2) and the percentage of fibers in each category is shown in the bar graph. b, Density of nuclei in muscle biopsies from control (gray circles) and FUS-ALS patients (green circles) as a proxy for muscle atrophy. The number of nuclei per field of 13,547 μm2 is shown in the dot plot. One-way ANOVA with Tukey’s multiple comparisons test; **p<0.01, ***p<0.001; N= 4 fields. Average ± SEM. c, Dot plot showing cytoplasmic FUS staining intensity in muscle biopsies from control (gray circles) and FUS-ALS patients (green circles). Kruskal-Wallis test with Dunn’s multiple comparisons test; *p<0.05, ***p<0.0001. Number of fields in which the average cytoplasmic FUS staining intensity is measured=3 (CNTL4), 5 (CNTL5), 2 (CNTL6), 4 (FUS1), 5 (FUS4), 10 (FUS5). Average ± SEM. d, Dot plot showing BTX-positive endplate area in muscle biopsies from control (gray circles) and FUS-ALS patients (green circles). One-way ANOVA with Tukey’s multiple comparisons test; *p<0.05, ***p<0.0001. Number of endplates=3 (CNTL4), 6 (CNTL5), 4 (CNTL6), 5 (FUS1), 7 (FUS4), 24 (FUS5). Average ± SEM.
Supplementary Figure 10 Working model illustrating how FUS may regulate transcription of Chrn genes in subsynaptic myonuclei.
Agrin secreted from motor nerve endings binds to LRP4 on postsynaptic membranes, resulting in MuSK activation and subsequent activation of ERM through Rac1 and JNK pathways. FUS is enriched in subsynaptic nuclei, interacts with ERM, binds to AChR gene promoters and cooperates with ERM to stimulate transcription of Chrn genes.
Supplementary information
Supplementary Information
Supplementary Figs. 1–11, Supplementary Tables 1–4 and Supplementary Note 1.
Rights and permissions
About this article
Cite this article
Picchiarelli, G., Demestre, M., Zuko, A. et al. FUS-mediated regulation of acetylcholine receptor transcription at neuromuscular junctions is compromised in amyotrophic lateral sclerosis. Nat Neurosci 22, 1793–1805 (2019). https://doi.org/10.1038/s41593-019-0498-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41593-019-0498-9
This article is cited by
-
Molecular hallmarks of ageing in amyotrophic lateral sclerosis
Cellular and Molecular Life Sciences (2024)
-
Generation of functional posterior spinal motor neurons from hPSCs-derived human spinal cord neural progenitor cells
Cell Regeneration (2023)
-
Genetics of amyotrophic lateral sclerosis: seeking therapeutic targets in the era of gene therapy
Journal of Human Genetics (2023)
-
Efficient generation of a self-organizing neuromuscular junction model from human pluripotent stem cells
Nature Communications (2023)
-
Post-synaptic specialization of the neuromuscular junction: junctional folds formation, function, and disorders
Cell & Bioscience (2022)