Abstract
Common risk factors for psychiatric and other brain disorders are likely to converge on biological pathways influencing the development and maintenance of brain structure and function across life. Using structural MRI data from 45,615 individuals aged 3–96 years, we demonstrate distinct patterns of apparent brain aging in several brain disorders and reveal genetic pleiotropy between apparent brain aging in healthy individuals and common brain disorders.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The raw data incorporated in this work were gathered from various resources. Material requests will need to be placed with individual principal investigators. A detailed overview of the included cohorts is provided in Supplementary Table 1. GWAS summary statistics for the brain age gaps as well as the models needed to predict brain age in independent cohorts are available at github.com/tobias-kaufmann.
Code availability
Code needed to run brain age prediction models is available at github.com/tobias-kaufmann (see Data availability). Additional R statistics53 code is available from the authors upon request.
Change history
17 December 2019
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
References
WHO. World Health Statistics 2016: monitoring health for the SDGs. https://www.who.int/gho/publications/world_health_statistics/2016/en/ (2016).
Insel, T. R. & Cuthbert, B. N. Brain disorders? Precisely. Science 348, 499–500 (2015).
Prince, M. et al. No health without mental health. Lancet 370, 859–877 (2007).
Parikshak, N. N., Gandal, M. J. & Geschwind, D. H. Systems biology and gene networks in neurodevelopmental and neurodegenerative disorders. Nat. Rev. Genet 16, 441–458 (2015).
Marin, O. Developmental timing and critical windows for the treatment of psychiatric disorders. Nat. Med 22, 1229–1238 (2016).
Insel, T. R. Rethinking schizophrenia. Nature 468, 187–193 (2010).
Aubert-Broche, B. et al. Onset of multiple sclerosis before adulthood leads to failure of age-expected brain growth. Neurology 83, 2140–2146 (2014).
Masters, C. L. et al. Alzheimer’s disease. Nat. Rev. Dis. Primers 1, 15056 (2015).
Dosenbach, N. U. et al. Prediction of individual brain maturity using fMRI. Science 329, 1358–1361 (2010).
Franke, K., Ziegler, G., Klöppel, S. & Gaser, C. & Alzheimera’s Disease Neuroimaging Initiative. Estimating the age of healthy subjects from T1-weighted MRI scans using kernel methods: exploring the influence of various parameters. Neuroimage 50, 883–892 (2010).
Cole, J. H. & Franke, K. Predicting age using neuroimaging: innovative brain ageing biomarkers. Trends Neurosci. 40, 681–690 (2017).
Ritchie, S. J. et al. Sex differences in the adult human brain: evidence from 5216 UK biobank participants. Cereb. Cortex 28, 2959–2975 (2018).
Cole, J. H. et al. Predicting brain age with deep learning from raw imaging data results in a reliable and heritable biomarker. Neuroimage 163, 115–124 (2017).
Bansal, V. et al. Genome-wide association study results for educational attainment aid in identifying genetic heterogeneity of schizophrenia. Nat. Commun. 9, 3078 (2018).
Ellison-Wright, I. & Bullmore, E. Anatomy of bipolar disorder and schizophrenia: a meta-analysis. Schizophr. Res. 117, 1–12 (2010).
Jernigan, T. L., Salmon, D. P., Butters, N. & Hesselink, J. R. Cerebral structure on MRI, part II: specific changes in Alzheimer’s and Huntington’s diseases. Biol. psychiatry 29, 68–81 (1991).
Wolfers, T. et al. Mapping the heterogeneous phenotype of schizophrenia and bipolar disorder using normative models. Jama Psychiat 75, 1146–1155 (2018).
Ecker, C., Bookheimer, S. Y. & Murphy, D. G. Neuroimaging in autism spectrum disorder: brain structure and function across the lifespan. Lancet Neurol. 14, 1121–1134 (2015).
Faraone, S. V. et al. Attention-deficit/hyperactivity disorder. Nat. Rev. Dis. Primers 1, 15020 (2015).
Andreassen, O. A. et al. Genetic pleiotropy between multiple sclerosis and schizophrenia but not bipolar disorder: differential involvement of immune-related gene loci. Mol. psychiatry 20, 207 (2015).
Fischl, B. et al. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron 33, 341–355 (2002).
Rosen, A. F. G. et al. Quantitative assessment of structural image quality. Neuroimage 169, 407–418 (2018).
Smith, S. M. & Nichols, T. E. Statistical challenges in “Big Data” human neuroimaging. Neuron 97, 263–268 (2018).
Glasser, M. F. et al. A multi-modal parcellation of human cerebral cortex. Nature 536, 171–178 (2016).
Ho, D., Imai, K., King, G. & Stuart, E. A. MatchIt: nonparametric preprocessing for parametric causal inference. J. Stat. Softw. 42, 1–28 (2011).
Chen, T. & Guestrin, C. XGBoost: a scalable tree boosting system. In Proc. 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining 785–794 (ACM, 2016).
Chen, T., et al. Xgboost: extreme gradient boosting. R package v0.4-2 https://cran.r-project.org/web/packages/xgboost/ (2015).
Le, T. T. et al. A nonlinear simulation framework supports adjusting for age when analyzing BrainAGE. Front. Aging Neurosci. 10, 317 (2018).
Zuber, V. & Strimmer, K. Care. R package v 1.1.10. https://cran.r-project.org/web/packages/care/care.pdf (2017).
Cole, J. H. et al. Brain age predicts mortality. Mol. psychiatry 23, 1385–1392 (2018).
Nakagawa, S. & Cuthill, I. C. Effect size, confidence interval and statistical significance: a practical guide for biologists. Biol. Rev. Camb. Philos. Soc. 82, 591–605 (2007).
Viechtbauer, W. Conducting meta-analysis in R with the metafor package. J. Stat. Softw. 36, 1–48 (2010).
Warnes, G. R. et al. R Package gplots: various R programming tools for plotting data. https://cran.r-project.org/web/packages/gplots/gplots.pdf (2016).
Gu, Z. R. Package circlize: circular visualization. https://cran.r-project.org/web/packages/circlize/circlize.pdf (2017).
Bycroft, C. et al. TheUK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Bulik-Sullivan, B. K. et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015).
Bulik-Sullivan, B. et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 47, 1236–1241 (2015).
Nichols, T., Brett, M., Andersson, J., Wager, T. & Poline, J.-B. Valid conjunction inference with the minimum statistic. Neuroimage 25, 653–660 (2005).
Andreassen, O. A. et al. Improved detection of common variants associated with schizophrenia by leveraging pleiotropy with cardiovascular-disease risk factors. Am. J. Hum. Genet. 92, 197–209 (2013).
Grove, J. et al. Identification of common genetic risk variants for autism spectrum disorder. Nat. Genet. 51, 431–444 (2019).
Demontis, D. et al. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nature Genet. 51, 63–75 (2018).
Schizophrenia Working Group of the PGC. et al. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421 (2014).
Stahl, E. A. et al. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat. Genet. 51, 793–803 (2019).
Patsopoulos, N. et al. The multiple sclerosis genomic map: role of peripheral immune cells and resident microglia in susceptibility. Preprint at biorXiv https://www.biorxiv.org/content/10.1101/143933v1 (2017).
Wray, N. R. et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat. Genet. 50, 668–681 (2018).
Lambert, J.-C. et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet. 45, 1452 (2013).
McLaren, W. et al. The ensembl variant effect predictor. Genome Biol. 17, 122 (2016).
Pedersen, G. & Karterud, S. The symptom and function dimensions of the Global Assessment of Functioning (GAF) scale. Compr. Psychiatry 53, 292–298 (2012).
Kay, S. R., Fiszbein, A. & Opfer, L. A. The positive and negative syndrome scale (PANSS) for schizophrenia. Schizophr. Bull. 13, 261 (1987).
Kurtzke, J. F. Rating neurologic impairment in multiple sclerosis: an expanded disability status scale (EDSS). Neurology 33, 1444–1444 (1983).
Folstein, M. F., Folstein, S. E. & McHugh, P. R. “Mini-mental state”: a practical method for grading the cognitive state of patients for the clinician. J. Psychiatr. Res. 12, 189–198 (1975).
R Core Team. R: A Language and Environment for Statistical Computing. (R Foundation for Statistical Computing, Vienna, Austria, 2013).
Acknowledgements
The author list between I.A. and M.Z. is in alphabetic order. The authors were funded by the Research Council of Norway (276082 LifespanHealth (T.K.), 213837 (O.A.A), 223273 NORMENT (O.A.A.), 204966 (L.T.W.), 229129 (O.A.A.), 249795 (L.T.W.), 273345 (L.T.W.) and 283798 SYNSCHIZ (O.A.A.)), the South-Eastern Norway Regional Health Authority (2013-123 (O.A.A.), 2014-097 (L.T.W.), 2015-073 (L.T.W.) and 2016083 (L.T.W.)), Stiftelsen Kristian Gerhard Jebsen (SKGJ-MED-008), the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (ERC Starting Grant, Grant agreement No. 802998 BRAINMINT (L.T.W.)), NVIDIA Corporation GPU Grant (T.K.), and the European Commission 7th Framework Programme (602450, IMAGEMEND (A.M.-L.)). The data used in this study were gathered from various sources. A detailed overview of the included cohorts and acknowledgement of their respective funding sources and cohort-specific details is provided in Supplementary Table 1. Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu), the AddNeuroMed consortium and the Pediatric Imaging, Neurocognition and Genetics (PING) study database (www.chd.ucsd.edu/research/ping-study.html, now shared through the NIMH Data Archive (NDA)). The investigators within the ADNI and PING studies contributed to the design and implementation of ADNI and PING or provided data but did not participate in the analysis or writing of this report. This publication is solely the responsibility of the authors and does not necessarily represent the views of the National Institutes of Health or PING investigators. Complete listings of participating sites and study investigators can be found at http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf and http://pingstudy.ucsd.edu/investigators.html. The AddNeuroMed consortium was led by S.L., B.V., P.M., M.T., I.K. and H.S.
Author information
Authors and Affiliations
Consortia
Contributions
T.K. and L.T.W. conceived the study, T.K., N.T.D. and L.T.W. pre-processed all data in Freesurfer, N.T.D., M.J.L., C.L.B, L.B.N., L.T.W. and T.K. performed quality control of the data, T.K. performed the analysis with contributions from L.T.W. and D.v.d.M., and T.K., L.T.W., N.T.D., D.v.d.M. and O.A.A. contributed to interpretation of the results. All remaining authors were involved in data collection at various sites as well as cohort-specific tasks. T.K. and L.T.W. wrote the first draft of the paper and all authors contributed to and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
Some authors received educational speaker’s honoraria from Lundbeck (O.A.A., A.B., T.E., M.Z., N.I.L.), Sunovion (O.A.A.), Shire (B.F.), Medice (B.F.), Otsuka (A.B., M.Z.), Janssen (A.B.), Roche (M.Z.), Ferrer (M.Z.), Trommsdorff (M.Z.) and Servier (M.Z.), all unrelated to this work. A.B. is a stockholder of Hoffmann-La Roche and has received consultant fees from Biogen Idec. S.L. is currently an employee of Janssen-Cilag, but contribution to this work was completed prior to this employment. E.A.H., E.G.C., M.K.B., P.S., and H.F.H. have received travel support, honoraria for advice and/or lecturing from Almirall (E.G.C.), Biogen Idec (E.G.C., H.F.H., M.K.B.), Sanofi-Genzyme (E.G.C., H.F.H., E.A.H.), Merck (E.G.C., H.F.H., E.A.H., P. S.), Novartis (E.G.C., H.F.H., M.K.B.), Roche (E.G.C., H.F.H.), Sanofi-Aventis (E.G.C., H.F.H.) and Teva (E.G.C., H.F.H.). E.G.C. anf H.F.H. have received unrestricted research grants from Novartis (E.G.C., H.F.H.), Biogen Idec (E.G.C.) and Sanofi-Genzyme (E.G.C.). G.P. has been the academic supervisor of a Roche collaboration grant (years 2015-2016) that funds his salary. None of the mentioned external parties had any role in the analysis, writing or decision to publish this work. All other authors declare no competing interests.
Additional information
Peer review information Nature Neuroscience thanks Janine Bijsterbosch, Gagan Wig, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1
Outline of the main analysis pipeline.
Supplementary Figure 2 Validation of the prediction models confirmed validity of the models.
(a) The Pearson correlation between chronological age and predicted brain age estimated using 5-fold cross-validation within the training set (n=35,474) for each feature set. (b) The Pearson correlation between chronological age and predicted brain age estimated in the ten independent test samples. The test samples with cases and matched controls comprised n=925 ASD / n=925 HC, n=725 ADHD / n=725 HC, n=94 SZRISK / n=94 HC, n=1110 SZ / n=1110 HC, n=300 PSYMIX / n=300 HC, n=459 BD / n=459 HC, n=254 MS / n=254 HC, n=208 MDD / n=208 HC, n=974 MCI / n=974 HC, n=739 DEM / n=739 HC; in total n=10,141 independent subjects.
Supplementary Figure 3 Comparison of prediction models in data from the UK Biobank.
Our main analysis used xgboost models for estimating brain age. The figure compares predicted brain age (a) and brain age gaps (b) from xgboost to shrinkage linear models (slm) and to a free public brain age estimation approach (brainageR). Predicted brain age and resulting brain age gaps were highly correlated between approaches (Pearson r; analysis performed in n=23,602 healthy individuals from the UK Biobank; two-sided; all P < FDR; Benjamini-Hochberg). All brain age gaps account for age, age², sex, scanning site and Euler number.
Supplementary Figure 4 Comparison of models trained within site to models trained across site.
In the test sample (n=10,141 independent subjects), for each diagnosis and scanner, we trained a machine learning model on data from all available healthy controls acquired at a given scanner and predicted brain age on data from all cases collected at the respective scanner. Predicted brain age in cases from the within-site models correlated significantly with predicted brain age from the main models (mid column). Likewise, the resulting brain age gaps (right column) were significantly correlated between within-site models (accounted for age, age², sex and Euler number) and main models (accounted for age, age², sex, Euler number and scanning site), indicating that scanning-site independent models provide similar estimates of apparent aging patterns as the models built on cross-site imaging data. The figure reports Pearson r with two-sided p values, all P<FDR (Benjamini-Hochberg). Model performance (left column) is higher in the main models that use more data (see also Supplementary Fig. 5), and we have therefore used the across-site predictions for the main analysis.
Supplementary Figure 5 Impact of sample size on brain age prediction model performance.
Forty random subsets of data from 100, 500, 1000, 2000, 5000, 10000, and 20000 individuals were drawn and corresponding models cross-validated. In addition, results from the full sample (n=35,474) is shown. With increasing sample size, performance of the models increased, with less variation across runs. The whiskers in the boxplot extend from the hinge to the largest/smallest value no further than 1.5 * IQR from the hinge.
Supplementary Figure 6 Case-control differences on principal components in the test sets.
This analysis quantifies variance in the features in relation to case-control differences. We conducted a PCA in each test sample, yielding components that capture the variance across the 1118 brain imaging features, and assessed case-control differences in models accounting for age, age², sex, scanning site and Euler number. Except for MDD and SZRISK, all test samples showed significant case-control differences (black boxes indicate p < FDR; Benjamini-Hochberg; two-sided). The test samples with cases and matched controls comprised n=925 ASD / n=925 HC, n=725 ADHD / n=725 HC, n=94 SZRISK / n=94 HC, n=1110 SZ / n=1110 HC, n=300 PSYMIX / n=300 HC, n=459 BD / n=459 HC, n=254 MS / n=254 HC, n=208 MDD / n=208 HC, n=974 MCI / n=974 HC, n=739 DEM / n=739 HC; in total n=10,141 independent subjects.
Supplementary Figure 7 Meta-analysis confirmed mega-analysis results.
Individual study sample sizes are depicted in Supplementary Table 2. All Cohen’s d effect sizes for the effect of group accounted for age, age², sex and Euler number. Further, Cohen’s d for all cohorts that were collected at multiple sites also accounted for scanning site. For each sample, the center of the square reflects the estimated effect size and the error bars depict the 95% confidence interval.
Supplementary Figure 8 Replication of results in a subset of 40,301 individuals following more stringent quality control.
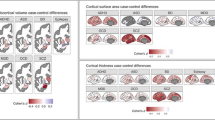
(a) Replication of group effects (for comparison, see Fig. 2a). The test samples in this sanity check analysis comprised n=770 ASD / n=770 HC, n=654 ADHD / n=654 HC, n=80 SZRISK / n=80 HC, n=931 SZ / n=931 HC, n=251 PSYMIX / n=251 HC, n=398 BD / n=398 HC, n=219 MS / n=219 HC, n=177 MDD / n=177 HC, n=803 MCI / n=803 HC, n=579 DEM / n=579 HC. Cohen’s d effect sizes (pooled standard deviation units) and two-sided P-values are provided. (b) Replication of spatial brain age gap patterns (for comparison, see Fig. 2b). Colours indicate Cohen’s d effect sizes for group comparisons. Sample size as specified in panel a. (c) Replication of interaction effect pattern (for comparison, see Fig. 2c). Sample size as specified in panel a yet excluding HC; n=4862 independent subjects. Only significant (p<FDR; Benjamini-Hochberg) effects are shown. (d) Replication of associations with clinical and cognitive scores (for comparison, see Fig. 2d). Associations were computed using linear models accounting for age, age², sex, scanning site and Euler number, and the resulting t-statistics were transformed to r. Black box indicates significance after correction for multiple comparison (p<FDR; Benjamini-Hochberg; two-sided). Sample size comprised n=340 SZ for GAFsymptom, n=234 SZ for GAFfunction, n=564 SZ for PANSSpositive, n=549 SZ for PANSSnegative, n=157 MS for EDSS, n=752 MCI and n=535 DEM for MMSE.
Supplementary Figure 9 Clustering of regional brain age gap patterns reveals disorder specific patterns.
The figure indicates hierarchical clustering of the Spearman correlation matrix of effect sizes from Fig. 2b (eight estimates of Cohen d per diagnosis (full brain, occipital, frontal, temporal, parietal, cingulate, insula, subcortical)).
Supplementary Figure 10 Results from 1260 repeated measures ANOVAs confirm group x region interaction effects in brain age patterns.
Strongest effects were observed between MS and SZ, MS and PSYMIX, MS and BD, MDD and MS, as well as DEM and SZ, suggesting divergent aging patterns in these disorders. The samples comprised n=925 ASD, n=725 ADHD, n=94 SZRISK, n=1110 SZ, n=300 PSYMIX, n=459 BD, n=254 MS, n=208 MDD, n=974 MCI, n=739 DEM; in total n=5788 independent subjects. Significant (p<FDR; Benjamini-Hochberg) effects are shown in white text colour.
Supplementary Figure 11 Scatter plots for associations with clinical and cognitive data in patient groups.
Subplots correspond to the associations reported in Fig. 2d. The analysis was performed in patient data only (excluding controls). Clinical data was available for SZ (GAFsymptom n=389, GAFfunction n=269, PANSSpositive n=646, PANSSnegative n=626), MS (EDSS n=195) and MCI/DEM (MMSE n=907 MCI and n=686 DEM). Shaded areas reflect confidence intervals. Associations were computed using linear models accounting for age, age², sex, scanning site and Euler number, and the resulting t-statistics were transformed to r. For assessment of significance, see Fig. 2d.
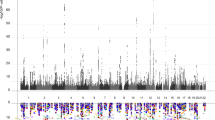
Supplementary Figure 12 Cross trait LD-score regression (Rg) between brain age gaps and psychiatric disorders.
GWAS on brain age gaps was performed in data from n=20,170 healthy adult individuals with European ancestry. One association (ADHD with cingulate brain age gap) survived multiple comparison testing (P<FDR; Benjamini-Hochberg, two-sided).
Supplementary Figure 13 Validation of the quality assurance pipeline.
A sample of 1521 individuals was manually quality controlled and data from individuals marked for in- or exclusion. This method was compared against Euler numbers (a-b) and against automated decisions based on data deceeding 3 (c) or 1 (d) standard deviation in Euler numbers. Together, these results confirm that Euler number is sensitive to data quality and can be used for automated quality control. We used 3 SD as the criterion for the main analysis, allowing us to exploit the full sample size at the risk of missing a proportion of outliers. To mitigate this risk, we also provide a re-analysis of the data in the supplement using 1 SD as the exclusion criterion. Panels a-b report t-statistics and two-sided P-values from one t-test per hemisphere. The whiskers in the boxplot extend from the hinge to the largest/smallest value no further than 1.5 * IQR from the hinge. The horizontal mid line depicts the median.
Supplementary information
Supplementary Figures 1–13 and Supplementary Tables 1 and 3
Supplementary Figures 1–13 and Supplementary Tables 1 and 3.
Supplementary Table 2
Summary of group size, age and sex for each cohort.
Supplementary Table 4
Sex specific brain age prediction models transferred well across sexes. In the training sample, 1008 out of 1118 features showed a significant sex difference (two-sided; Bonferroni level p<4e-05) in models accounting for age, age², scanning site and Euler number. Due to this, and also due to the possibility of subtle sex differences in age trajectories and mechanisms of brain aging (for example, related to hormones, etc.), we modeled male and female brains separately for predicting brain age in the main analysis. To investigate if lifespan changes in the brain are similar between sexes, we compared the prediction models. Age of all test participants predicted using the male model correlated strongly with their age predicted using the female model (Pearson r=0.99). In addition, the table illustrates the correlation (Pearson r) of predicted brain age with chronological age for different combinations of training models and test data. Together, these results suggest that the incorporated imaging features follow similar age trajectories across sexes.
Supplementary Table 5
Assessment of age by diagnosis interaction in the brain age gaps of ASD (n=925 ASD, n=925 HC) and ADHD (n=725 ADHD, n=725 HC). The linear models accounted for age, age², sex, scanning site and Euler number. None of the interaction effects survived correction for multiple comparison (FDR; Benjamini-Hochberg; two-sided).
Supplementary Table 6
Significant loci from conjunctional FDR analysis reflecting overlap between brain age gaps and the respective disorders. The gene column reflects the gene closest to the significant SNP as identified via the Ensembl Variable Effect Predictor48, unless the closest gene is more than 5000 bp away in which case no annotation is provided.
Rights and permissions
About this article
Cite this article
Kaufmann, T., van der Meer, D., Doan, N.T. et al. Common brain disorders are associated with heritable patterns of apparent aging of the brain. Nat Neurosci 22, 1617–1623 (2019). https://doi.org/10.1038/s41593-019-0471-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41593-019-0471-7
This article is cited by
-
Examining the reliability of brain age algorithms under varying degrees of participant motion
Brain Informatics (2024)
-
Menarche, pubertal timing and the brain: female-specific patterns of brain maturation beyond age-related development
Biology of Sex Differences (2024)
-
Genetic and phenotypic similarity across major psychiatric disorders: a systematic review and quantitative assessment
Translational Psychiatry (2024)
-
The genetic architecture of multimodal human brain age
Nature Communications (2024)
-
Brain asymmetries from mid- to late life and hemispheric brain age
Nature Communications (2024)