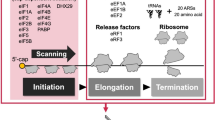
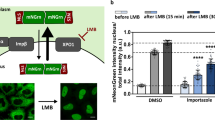
Abstract
Nucleotide repeat expansions in the C9orf72 gene are the most common cause of amyotrophic lateral sclerosis and frontotemporal dementia. Unconventional translation (RAN translation) of C9orf72 repeats generates dipeptide repeat proteins that can cause neurodegeneration. We performed a genetic screen for regulators of RAN translation and identified small ribosomal protein subunit 25 (RPS25), presenting a potential therapeutic target for C9orf72-related amyotrophic lateral sclerosis and frontotemporal dementia and other neurodegenerative diseases caused by nucleotide repeat expansions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others

Data availability
The data that support the findings of the present study are available from the corresponding author upon request.
References
Renton, A. E. et al. Neuron 72, 257–268 (2011).
DeJesus-Hernandez, M. et al. Neuron 72, 245–256 (2011).
Mori, K. et al. Science 339, 1335–1338 (2013).
Ash, P. E. et al. Neuron 77, 639–646 (2013).
Zu, T. et al. Proc. Natl Acad. Sci. USA 110, E4968–E4977 (2013).
Gendron, T. F. et al. Acta Neuropathol. 126, 829–844 (2013).
Gao, F. B., Richter, J. D. & Cleveland, D. W. Cell 171, 994–1000 (2017).
Cheng, W. et al. Nat. Commun. 9, 51 (2018).
Green, K. M. et al. Nat. Commun. 8, 2005 (2017).
Tabet, R. et al. Nat. Commun. 9, 152 (2018).
Landry, D. M., Hertz, M. I. & Thompson, S. R. Genes Dev. 23, 2753–2764 (2009).
Fuchs, G. et al. Proc. Natl Acad. Sci. USA 112, 319–325 (2015).
Hertz, M. I., Landry, D. M., Willis, A. E., Luo, G. & Thompson, S. R. Mol. Cell Biol. 33, 1016–1026 (2013).
Nishiyama, T., Yamamoto, H., Uchiumi, T. & Nakashima, N. Nucleic Acids Res. 35, 1514–1521 (2007).
Shi, Y. et al. Oncogene 35, 1015–1024 (2016).
Thandapani, P., O’Connor, T. R., Bailey, T. L. & Richard, S. Mol. Cell 50, 613–623 (2013).
Mizielinska, S. et al. Science 345, 1192–1194 (2014).
Shi, Y. et al. Nat. Med. 24, 313–325 (2018).
Cleary, J. D. & Ranum, L. P. Curr. Opin. Genet. Dev. 44, 125–134 (2017).
Green, K. M., Linsalata, A. E. & Todd, P. K. Brain Res. 1647, 30–42 (2016).
Kramer, N. J. et al. Science 353, 708–712 (2016).
Alberti, S., Gitler, A. D. & Lindquist, S. Yeast 24, 913–919 (2007).
Cooper, A. A. et al. Science 313, 324–328 (2006).
Gietz, R. D. & Schiestl, R. H. Nat. Protoc. 2, 38–41 (2007).
Gendron, T. F. et al. Sci. Transl. Med. 9, eaai7866 (2017).
Gendron, T. F. et al. Acta Neuropathol. 130, 559–573 (2015).
Kramer, N. J. et al. Nat. Genet. 50, 603–612 (2018).
Su, Z. et al. Neuron 83, 1043–1050 (2014).
Simone, R. et al. EMBO Mol. Med. 10, 22–31 (2017).
Banez-Coronel, M. et al. Neuron 88, 667–677 (2015).
Osterwalder, T., Yoon, K. S., White, B. H. & Keshishian, H. Proc. Natl Acad. Sci. USA 98, 12596–12601 (2001).
Son, E. Y. et al. Cell Stem Cell 9, 205–218 (2011).
Acknowledgements
This work was supported by National Institutes of Health (NIH) grants nos. R35NS097263 (A.D.G.), AI099506 (J.D.P), AG064690 (A.D.G. and J.D.P.), R35NS097273 (L.P.), P01NS099114 (T.F.G., L.P) and R01NS097850 (J.K.I.), the Robert Packard Center for ALS Research at Johns Hopkins (L.P., A.D.G.), Target ALS (L.P., A.D.G., T.F.G.), the US Department of Defense (J.D.P., A.D.G., J.K.I.), the Muscular Dystrophy Association (J.K.I.), 2T32HG000044-21 NIHGRI training grant (S.B.Y.), the Brain Rejuvenation Project of the Wu Tsai Neurosciences Institute (A.D.G.), the European Research Council grant no. ERC-2014-CoG-648716 (A.M.I.), Alzheimer’s Research UK (A.M.I.) and the Medical Research Council (A.M.I.). J.K.I. is supported by funding for a New York Stem Cell Foundation-Robertson Investigatorship. We thank Dr. L. Ranum (University of Florida) for sharing the huntingtin poly(alanine) antibody. We thank M. Bassik for the HeLa Cas9–BFP cell lines. We thank H. Tricoire (French National Center for Scientific Research) for the generous gift of the original elavGS 301.2 line.
Author information
Authors and Affiliations
Contributions
This work was performed and the paper written by S.B.Y. under the mentorship of A.D.G. T.F.G. contributed ELISA assays to detect RAN peptides and analyses under the mentorship of L.P. T.N., I.G. and A.T. contributed Drosophila studies under the mentorship of L.P. and A.M.I. N.R.G. contributed to polysome profiling studies and analyses, under the mentorship of M.B. R.G. contributed to RAN translation studies and analyses, under the mentorship of J.D.P. Y.S. and G.R. contributed induced motor neuron studies and analyses, under the mentorship of J.K.I. N.J.K. contributed to studies of ATXN2 RAN translation. L.N., S.F. and T.J.I.D. contributed to studies of C9orf72 RAN translation.
Corresponding author
Ethics declarations
Competing interests
A.D.G. has served as a consultant for Aquinnah Pharmaceuticals, Prevail Therapeutics and Third Rock Ventures and is a scientific founder of Maze Therapeutics.
Additional information
Peer review information: Nature Neuroscience thanks D. Dormann and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Yamada, S.B., Gendron, T.F., Niccoli, T. et al. RPS25 is required for efficient RAN translation of C9orf72 and other neurodegenerative disease-associated nucleotide repeats. Nat Neurosci 22, 1383–1388 (2019). https://doi.org/10.1038/s41593-019-0455-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41593-019-0455-7
This article is cited by
-
Translation dysregulation in neurodegenerative diseases: a focus on ALS
Molecular Neurodegeneration (2023)
-
rDNA Transcription in Developmental Diseases and Stem Cells
Stem Cell Reviews and Reports (2023)
-
Identification of potential biomarkers for pathogenesis of Alzheimer’s disease
Hereditas (2021)
-
Slc1a3-2A-CreERT2 mice reveal unique features of Bergmann glia and augment a growing collection of Cre drivers and effectors in the 129S4 genetic background
Scientific Reports (2021)
-
A C. elegans model of C9orf72-associated ALS/FTD uncovers a conserved role for eIF2D in RAN translation
Nature Communications (2021)