Abstract
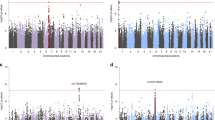
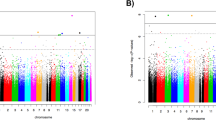
Post-traumatic stress disorder (PTSD) is a major problem among military veterans and civilians alike, yet its pathophysiology remains poorly understood. We performed a genome-wide association study and bioinformatic analyses, which included 146,660 European Americans and 19,983 African Americans in the US Million Veteran Program, to identify genetic risk factors relevant to intrusive reexperiencing of trauma, which is the most characteristic symptom cluster of PTSD. In European Americans, eight distinct significant regions were identified. Three regions had values of P < 5 × 10−10: CAMKV; chromosome 17 closest to KANSL1, but within a large high linkage disequilibrium region that also includes CRHR1; and TCF4. Associations were enriched with respect to the transcriptomic profiles of striatal medium spiny neurons. No significant associations were observed in the African American cohort of the sample. Results in European Americans were replicated in the UK Biobank data. These results provide new insights into the biology of PTSD in a well-powered genome-wide association study.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The GWAS summary statistics generated during and/or analyzed during the current study are available via dbGAP; the dbGaP accession assigned to the Million Veteran Program is phs001672.v1.p. The website is: https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001672.v1.p1. Additionally, the data that support the findings of this study are available from the corresponding authors upon reasonable request.
References
Fulton, J. J. et al. The prevalence of posttraumatic stress disorder in operation enduring freedom/operation iraqi freedom (OEF/OIF) veterans: a meta-analysis. J. Anxiety Disord. 31, 98–107 (2015).
Logue, M. W. et al. A genome-wide association study of post-traumatic stress disorder identifies the retinoid-related orphan receptor alpha (RORA) gene as a significant risk locus. Mol. Psychiatry 18, 937–942 (2013).
Stein, M. B. et al. Genome-wide association studies of posttraumatic stress disorder in 2 cohorts of US army soldiers. JAMA Psychiatry 73, 695–704 (2016).
Xie, P. et al. Genome-wide association study identifies new susceptibility loci for posttraumatic stress disorder. Biol. Psychiatry 74, 656–663 (2013).
Duncan, L. E. et al. Largest GWAS of PTSD (N=20 070) yields genetic overlap with schizophrenia and sex differences in heritability. Mol. Psychiatry 23, 666–673 (2018).
Blanchard, E. B., Jones-Alexander, J., Buckley, T. C. & Forneris, C. A. Psychometric properties of the PTSD Checklist (PCL). Behav. Res. Ther. 34, 669–673 (1996).
Battle, A., Brown, C. D., Engelhardt, B. E. & Montgomery, S. B. Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017).
GTEx Consortium. Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017).
Skene, N. G. et al. Genetic identification of brain cell types underlying schizophrenia. Nat. Genet. 50, 825–833 (2018).
Barbano, A. C. et al. Clinical implications of the proposed ICD-11 PTSD diagnostic criteria. Psychol. Med. 49, 483–490 (2019).
Stefansson, H. et al. A common inversion under selection in Europeans. Nat. Genet. 37, 129–137 (2005).
Cáceres, A., Sindi, S. S., Raphael, B. J., Cáceres, M. & González, J. R. Identification of polymorphic inversions from genotypes. BMC Bioinformatics 13, 28 (2012).
Amstadter, A. B. et al. Corticotrophin-releasing hormone type 1 receptor gene (CRHR1) variants predict posttraumatic stress disorder onset and course in pediatric injury patients. Dis. Markers 30, 89–99 (2011).
Kasckow, J. W., Baker, D. & Geracioti, T. D. Jr. Corticotropin-releasing hormone in depression and post-traumatic stress disorder. Peptides 22, 845–851 (2001).
McFarlane, A. C., Barton, C. A., Yehuda, R. & Wittert, G. Cortisol response to acute trauma and risk of posttraumatic stress disorder. Psychoneuroendocrinology 36, 720–727 (2011).
Smith, D. J. et al. Genome-wide analysis of over 106 000 individuals identifies 9 neuroticism-associated loci. Mol. Psychiatry 21, 749–757 (2016).
Stefansson, H. et al. Common variants conferring risk of schizophrenia. Nature 460, 744–747 (2009).
Cross-Disorder Group of the Psychiatric Genomics Consortium. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet 381, 1371–1379 (2013).
Ruderfer, D. M.et al. Polygenic dissection of diagnosis and clinical dimensions of bipolar disorder and schizophrenia. Mol. Psychiatry 19, 1017–1024 (2014).
McGrath, J. J. et al. Trauma and psychotic experiences: transnational data from the world mental health survey. Br. J. Psychiatry 211, 373–380 (2017).
Waters, F., Blom, J. D., Jardri, R., Hugdahl, K. & Sommer, I. E. C. Auditory hallucinations, not necessarily a hallmark of psychotic disorder. Psychol. Med. 48, 529–536 (2018).
Ravindran, L. N. & Stein, M. B. The pharmacologic treatment of anxiety disorders: a review of progress. J. Clin. Psychiatry 71, 839–854 (2010).
Krystal, J. H. et al. Adjunctive risperidone treatment for antidepressant-resistant symptoms of chronic military service-related PTSD: a randomized trial. JAMA 306, 493–502 (2011).
Gelernter, J. et al. Genome-wide association study of opioid dependence: multiple associations mapped to calcium and potassium pathways. Biol. Psychiatry 76, 66–74 (2014).
Okbay, A. et al. Genome-wide association study identifies 74 loci associated with educational attainment. Nature 533, 539–542 (2016).
Alexander, M. et al. Rab27-dependent exosome production inhibits chronic inflammation and enables acute responses to inflammatory stimuli. J. Immunol. 199, 3559–3570 (2017).
Michopoulos, V., Powers, A., Gillespie, C. F., Ressler, K. J. & Jovanovic, T. Inflammation in fear- and anxiety-based disorders: PTSD, GAD, and beyond. Neuropsychopharmacology 42, 254–270 (2017).
Howard, D. M. et al. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat. Neurosci. 22, 343–352 (2019).
Sumner, J. A. et al. Post-traumatic stress disorder symptoms and risk of hypertension over 22 years in a large cohort of younger and middle-aged women. Psychol. Med. 46, 3105–3116 (2016).
Roy, S. S., Foraker, R. E., Girton, R. A. & Mansfield, A. J. Posttraumatic stress disorder and incident heart failure among a community-based sample of US veterans. Am. J. Public Health 105, 757–763 (2015).
Raskind, M. A. et al. Trial of prazosin for post-traumatic stress disorder in military veterans. N. Engl. J. Med. 378, 507–517 (2018).
Gaziano, J. M. et al. Million Veteran Program: a mega-biobank to study genetic influences on health and disease. J. Clin. Epidemiol. 70, 214–223 (2016).
Wilkins, K. C., Lang, A. J. & Norman, S. B. Synthesis of the psychometric properties of the PTSD checklist (PCL) military, civilian, and specific versions. Depress. Anxiety 28, 596–606 (2011).
Das, S. et al. Next-generation genotype imputation service and methods. Nat. Genet. 48, 1284–1287 (2016).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Abraham, G. & Inouye, M. Fast principal component analysis of large-scale genome-wide data. PLoS ONE 9, e93766 (2014).
Auton, A. et al. A global reference for human genetic variation. Nature 526, 68–74 (2015).
Zhan, X., Hu, Y., Li, B., Abecasis, G. R. & Liu, D. J. RVTESTS: an efficient and comprehensive tool for rare variant association analysis using sequence data. Bioinformatics 32, 1423–1426 (2016).
de Leeuw, C. A., Mooij, J. M., Heskes, T. & Posthuma, D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 11, e1004219 (2015).
Watanabe, K., Taskesen, E., van Bochoven, A. & Posthuma, D. FUMA: functional mapping and annotation of genetic associations. Preprint at: https://doi.org/10.1101/110023 (2017).
Bulik-Sullivan, B. et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 47, 1236–1241 (2015).
Bulik-Sullivan, B. K., Loh, P. R., Finucane, H. K., Ripke, S. & Yang, J. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015).
Zheng, J. et al. HAPRAP: a haplotype-based iterative method for statistical fine mapping using GWAS summary statistics. Bioinformatics 33, 79–86 (2017).
Zheng, J. et al. LD hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics 33, 272–279 (2017).
Bycroft, C. et al. Genome-wide genetic data on ~500,000 UK Biobank participants. Preprint at https://doi.org/10.1101/166298 (2017).
Aulchenko, Y. S., Ripke, S., Isaacs, A. & van Duijn, C. M. GenABEL: an R library for genome-wide association analysis. Bioinformatics 23, 1294–1296 (2007).
Finucane, H. K. et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 47, 1228–1235 (2015).
Finucane, H. K. et al. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat. Genet. 50, 621–629 (2018).
Szklarczyk, D. et al. The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 45, D362–D368 (2017).
Euesden, J., Lewis, C. M. & O’Reilly, P. F. PRSice: polygenic risk score software. Bioinformatics 31, 1466–1468 (2015).
Acknowledgements
This research is based on data from the Million Veteran Program (MVP), Office of Research and Development, Veterans Health Administration, and was supported by the MVP and the VA Cooperative Studies Program (CSP) study no. 575B. The views expressed in this article are those of the authors and do not necessarily reflect the position or policy of the Department of Veterans Affairs or the United States government.
Author information
Authors and Affiliations
Consortia
Contributions
Genotyping effort: S.P., Y.S. and H.H.-Z. Analyses team: N.S., J.B., Q.L., Y.H., B.L., R. Polimanti, Q.C. and D.F.L. CSP575B team (database, phenotype and analytic efforts): K.R., M.A., K.H.C., Y.L., N.R., F.S., K.H., K.C., R.Q. and R. Pietrzak. MVP leadership (overall MVP design and supervision): J.M.G. and J.C. Analysis design: R. Polimanti, P.F.S., H.Z., J.G. and M.B.S. Project design and project leadership: J.G., J.C. and M.B.S. Writing leads: J.G. and M.B.S.
Corresponding author
Ethics declarations
Competing interests
J.G. is named as a co-inventor on PCT patent application no. 15/878,640 entitled: “Genotype-guided dosing of opioid agonists,” filed 24 January 2018. M.B.S. has in the past 3 years been a consultant for Actelion, Aptinyx, Bionomics, Dart Neuroscience, Healthcare Management Technologies, Janssen, Jazz Pharmaceuticals, Neurocrine Biosciences, Oxeia Biopharmaceuticals, Pfizer, and Resilience Therapeutics. M.B.S. owns founders shares and stock options in Resilience Therapeutics and has stock options in Oxeia Biopharmaceuticals.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Table 3
Top 10,000 variants in EAs (LD pruned)
Supplementary Table 4
Top 10,000 variants in AAs (LD pruned)
Rights and permissions
About this article
Cite this article
Gelernter, J., Sun, N., Polimanti, R. et al. Genome-wide association study of post-traumatic stress disorder reexperiencing symptoms in >165,000 US veterans. Nat Neurosci 22, 1394–1401 (2019). https://doi.org/10.1038/s41593-019-0447-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41593-019-0447-7
This article is cited by
-
Effects of sex and gender on the etiologies and presentation of select internalizing psychopathologies
Translational Psychiatry (2024)
-
Fear memory regulation by the cAMP signaling pathway as an index of reexperiencing symptoms in posttraumatic stress disorder
Molecular Psychiatry (2024)
-
Genome-wide association analyses identify 95 risk loci and provide insights into the neurobiology of post-traumatic stress disorder
Nature Genetics (2024)
-
Genome-wide analysis of a model-derived binge eating disorder phenotype identifies risk loci and implicates iron metabolism
Nature Genetics (2023)
-
Genomic risk for post-traumatic stress disorder in families densely affected with alcohol use disorders
Molecular Psychiatry (2023)