Abstract
Multiple sclerosis (MS) is characterized by inflammatory insults that drive neuroaxonal injury. However, knowledge about neuron-intrinsic responses to inflammation is limited. By leveraging neuron-specific messenger RNA profiling, we found that neuroinflammation leads to induction and toxic accumulation of the synaptic protein bassoon (Bsn) in the neuronal somata of mice and patients with MS. Neuronal overexpression of Bsn in flies resulted in reduction of lifespan, while genetic disruption of Bsn protected mice from inflammation-induced neuroaxonal injury. Notably, pharmacological proteasome activation boosted the clearance of accumulated Bsn and enhanced neuronal survival. Our study demonstrates that neuroinflammation initiates toxic protein accumulation in neuronal somata and advocates proteasome activation as a potential remedy.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
Data generated for this study are available through the Gene Expression Omnibus under accession number GSE104899.
Code availability
The routine for the single-cell data analysis can be assessed as an R Notebook file via github (github.com/INIMS/Bsn_scRNA-seq).
References
Dendrou, C. A., Fugger, L. & Friese, M. A. Immunopathology of multiple sclerosis. Nat. Rev. Immunol. 15, 545–558 (2015).
Friese, M. A., Schattling, B. & Fugger, L. Mechanisms of neurodegeneration and axonal dysfunction in multiple sclerosis. Nat. Rev. Neurol. 10, 225–238 (2014).
Campbell, G. R. et al. Mitochondrial DNA deletions and neurodegeneration in multiple sclerosis. Ann. Neurol. 69, 481–492 (2011).
Forte, M. et al. Cyclophilin D inactivation protects axons in experimental autoimmune encephalomyelitis, an animal model of multiple sclerosis. Proc. Natl Acad. Sci. USA 104, 7558–7563 (2007).
Weisfeld-Adams, J. D., Katz Sand, I. B., Honce, J. M. & Lublin, F. D. Differential diagnosis of Mendelian and mitochondrial disorders in patients with suspected multiple sclerosis. Brain 138, 517–539 (2015).
Schattling, B. et al. TRPM4 cation channel mediates axonal and neuronal degeneration in experimental autoimmune encephalomyelitis and multiple sclerosis. Nat. Med. 18, 1805–1811 (2012).
Friese, M. A. et al. Acid-sensing ion channel-1 contributes to axonal degeneration in autoimmune inflammation of the central nervous system. Nat. Med. 13, 1483–1489 (2007).
Schattling, B. et al. Activity of NaV1.2 promotes neurodegeneration in an animal model of multiple sclerosis. JCI Insight 1, e89810 (2016).
Bading, H. Nuclear calcium signalling in the regulation of brain function. Nat. Rev. Neurosci. 14, 593–608 (2013).
Raddatz, B. B. et al. Transcriptomic meta-analysis of multiple sclerosis and its experimental models. PLoS One 9, e86643 (2014).
Heiman, M. et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell 135, 738–748 (2008).
Schmidt, E. F. et al. Identification of the cortical neurons that mediate antidepressant responses. Cell 149, 1152–1163 (2012).
Li, K., Nakajima, M., Ibañez-Tallon, I. & Heintz, N. A cortical circuit for sexually dimorphic oxytocin-dependent anxiety behaviors. Cell 167, 60–72.e11 (2016).
Thomson, S. R. et al. Cell-type-specific translation profiling reveals a novel strategy for treating fragile X syndrome. Neuron 95, 550–563.e5 (2017).
Sun, S. et al. Translational profiling identifies a cascade of damage initiated in motor neurons and spreading to glia in mutant SOD1-mediated ALS. Proc. Natl Acad. Sci. USA 112, E6993–E7002 (2015).
Brichta, L. et al. Identification of neurodegenerative factors using translatome-regulatory network analysis. Nat. Neurosci. 18, 1325–1333 (2015).
Chiti, F. & Dobson, C. M. Protein misfolding, amyloid formation, and human disease: a summary of progress over the last decade. Annu. Rev. Biochem. 86, 27–68 (2017).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Cho, H. et al. Differential innate immune response programs in neuronal subtypes determine susceptibility to infection in the brain by positive-stranded RNA viruses. Nat. Med. 19, 458–464 (2013).
Merico, D., Isserlin, R., Stueker, O., Emili, A. & Bader, G. D. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One 5, e13984 (2010).
Han, M. H. et al. Janus-like opposing roles of CD47 in autoimmune brain inflammation in humans and mice. J. Exp. Med. 209, 1325–1334 (2012).
Altrock, W. D. et al. Functional inactivation of a fraction of excitatory synapses in mice deficient for the active zone protein Bassoon. Neuron 37, 787–800 (2003).
Gundelfinger, E. D., Reissner, C. & Garner, C. C. Role of Bassoon and Piccolo in assembly and molecular organization of the active zone. Front. Synaptic Neurosci. 7, 19 (2016).
Vucetic, S., Brown, C. J., Dunker, A. K. & Obradovic, Z. Flavors of protein disorder. Proteins 52, 573–584 (2003).
Macosko, E. Z. et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015).
Trapnell, C. et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014).
Hetz, C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat. Rev. Mol. Cell Biol. 13, 89–102 (2012).
Bruckner, J. J. et al. Fife, a Drosophila Piccolo-RIM homolog, promotes active zone organization and neurotransmitter release. J. Neurosci. 32, 17048–17058 (2012).
Annamneedi, A. et al. Ablation of the presynaptic organizer Bassoon in excitatory neurons retards dentate gyrus maturation and enhances learning performance. Brain Struct. Funct. 223, 3423–3445 (2018).
Lee, B. H. et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature 467, 179–184 (2010).
Min, J. W., Lü, L., Freeling, J. L., Martin, D. S. & Wang, H. USP14 inhibitor attenuates cerebral ischemia/reperfusion-induced neuronal injury in mice. J. Neurochem. 140, 826–833 (2017).
Na, C. H. et al. Synaptic protein ubiquitination in rat brain revealed by antibody-based ubiquitome analysis. J. Proteome Res. 11, 4722–4732 (2012).
Menezes, M. J. et al. Mutation in mitochondrial ribosomal protein S7 (MRPS7) causes congenital sensorineural deafness, progressive hepatic and renal failure and lactic acidemia. Hum. Mol. Genet. 24, 2297–2307 (2015).
Nikić, I. et al. A reversible form of axon damage in experimental autoimmune encephalomyelitis and multiple sclerosis. Nat. Med. 17, 495–499 (2011).
Popovic, D., Vucic, D. & Dikic, I. Ubiquitination in disease pathogenesis and treatment. Nat. Med. 20, 1242–1253 (2014).
Lim, J. & Yue, Z. Neuronal aggregates: formation, clearance, and spreading. Dev. Cell 32, 491–501 (2015).
David, M. A. & Tayebi, M. Detection of protein aggregates in brain and cerebrospinal fluid derived from multiple sclerosis patients. Front. Neurol. 5, 251 (2014).
Gao, H. M. et al. Neuroinflammation and α-synuclein dysfunction potentiate each other, driving chronic progression of neurodegeneration in a mouse model of Parkinson’s disease. Environ. Health Perspect. 119, 807–814 (2011).
Bhaskar, K. et al. Regulation of tau pathology by the microglial fractalkine receptor. Neuron 68, 19–31 (2010).
Jarosz, D. F. & Khurana, V. Specification of physiologic and disease states by distinct proteins and protein conformations. Cell 171, 1001–1014 (2017).
Yabe, I. et al. Mutations in bassoon in individuals with familial and sporadic progressive supranuclear palsy-like syndrome. Sci. Rep. 8, 819 (2018).
Dresbach, T. et al. Assembly of active zone precursor vesicles: obligatory trafficking of presynaptic cytomatrix proteins Bassoon and Piccolo via a trans-Golgi compartment. J. Biol. Chem. 281, 6038–6047 (2006).
Waites, C. L. et al. Bassoon and Piccolo maintain synapse integrity by regulating protein ubiquitination and degradation. EMBO J. 32, 954–969 (2013).
Okerlund, N. D. et al. Bassoon controls presynaptic autophagy through Atg5. Neuron 93, 897–913.e7 (2017).
Sevigny, J. et al. The antibody aducanumab reduces Aβ plaques in Alzheimer’s disease. Nature 537, 50–56 (2016).
Harrigan, J. A., Jacq, X., Martin, N. M. & Jackson, S. P. Deubiquitylating enzymes and drug discovery: emerging opportunities. Nat. Rev. Drug Discov. 17, 57–78 (2018).
Walsh, D. M., Tseng, B. P., Rydel, R. E., Podlisny, M. B. & Selkoe, D. J. The oligomerization of amyloid β-protein begins intracellularly in cells derived from human brain. Biochemistry 39, 10831–10839 (2000).
Sperling, R., Mormino, E. & Johnson, K. The evolution of preclinical Alzheimer’s disease: implications for prevention trials. Neuron 84, 608–622 (2014).
Jucker, M. & Walker, L. C. Self-propagation of pathogenic protein aggregates in neurodegenerative diseases. Nature 501, 45–51 (2013).
Ontaneda, D., Thompson, A. J., Fox, R. J. & Cohen, J. A. Progressive multiple sclerosis: prospects for disease therapy, repair, and restoration of function. Lancet 389, 1357–1366 (2017).
Doyle, J. P. et al. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell 135, 749–762 (2008).
Heiman, M., Kulicke, R., Fenster, R. J., Greengard, P. & Heintz, N. Cell type-specific mRNA purification by translating ribosome affinity purification (TRAP). Nat. Protoc. 9, 1282–1291 (2014).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Durinck, S., Spellman, P. T., Birney, E. & Huber, W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 4, 1184–1191 (2009).
Cahoy, J. D. et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J. Neurosci. 28, 264–278 (2008).
Chen, J., Bardes, E. E., Aronow, B. J. & Jegga, A. G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 37, W305–W311 (2009).
Peng, K., Radivojac, P., Vucetic, S., Dunker, A. K. & Obradovic, Z. Length-dependent prediction of protein intrinsic disorder. BMC Bioinformatics 7, 208 (2006).
Dresbach, T. et al. Functional regions of the presynaptic cytomatrix protein bassoon: significance for synaptic targeting and cytomatrix anchoring. Mol. Cell. Neurosci. 23, 279–291 (2003).
Chien, C. L., Lu, K. S., Lin, Y. S., Hsieh, C. J. & Hirokawa, N. The functional cooperation of MAP1A heavy chain and light chain 2 in the binding of microtubules. Exp. Cell Res. 308, 446–458 (2005).
Kowalczyk, M. S. et al. Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Res. 25, 1860–1872 (2015).
Han, C., Jan, L. Y. & Jan, Y. N. Enhancer-driven membrane markers for analysis of nonautonomous mechanisms reveal neuron-glia interactions in Drosophila. Proc. Natl Acad. Sci. USA 108, 9673–9678 (2011).
Sun, Y. et al. Aging studies in Drosophila melanogaster. Methods Mol. Biol. 1048, 77–93 (2013).
Acknowledgements
We thank the UKE Mouse Pathology Facility for histopathology of EAE mice, N. Heintz for the Chat-bacTRAP mice, M. Heiman for advice on TRAP optimization, T. Dresbach for providing the Bsn DNA expression constructs, C.-L. Chien for providing the Map1a DNA expression construct and M. Binder and N. Akyüz for sequencing support. This work is supported by a Deutsche Forschungsgemeinschaft grant (grant no. FR 1720/11-1) and Oppenheim Förderpreis für Multiple Sklerose (Novartis) to M.A.F., as well as the Else Kröner-Fresenius-Stiftung (grant no. 2013_A217) to M.A.F. and K.E.D. D.M. is supported by the Swiss National Science Foundation (grant no. PP00P3_152928), Helmut Horten Foundation and Gebert-Rüf Foundation (grant no. GRS-049/13). U.T. and E.D.G. are supported by the Deutsche Forschungsgemeinschaft (grant no. CRC 854/B08).
Author information
Authors and Affiliations
Contributions
C.V. conducted the initial TRAP screening experiment. K.K.M. and K.E.D. helped optimizing the TRAP method. G.S. performed the RNA-seq. J.B.E. carried out the bioinformatics analysis. B.S., I.W., A.B. and S.C.R. performed the candidate validation by TRAP and histology. W.B. and D.M. conducted the human histopathology experiments. N.R. established the culture and sorting of N2a cells overexpressing Bsn. M.K. and J.B.E. performed the single-cell RNA-seq analysis. A.F., U.T. and E.D.G. generated the transgenic Bsn mice and flies and provided reagents and expertize. M.S.W., M.P. and P.S. performed the Drosophila experiments. B.S. performed EAE in Bsn knockout animals and together with J.B.E. in IU1-treated animals. B.S., S.T. and S.B. conducted the histopathological analysis in EAE. J.B.E., B.S. and M.A.F. designed the experiments and analyzed the data. J.B.E., B.S. and M.A.F. wrote the manuscript. M.A.F. conceived and supervised the study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Journal peer review information Nature Neuroscience thanks Joseph Dougherty, Hartmut Wekerle and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Fig. 1 Chat-L10a-eGFP expression validation and sample similarity analysis.
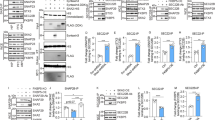
a, Representative immunohistochemical stainings of astrocytes (GFAP), vessels (CD31), microglia (Iba1), immune cells (CD45), oligodendrocytes (CNPase) and neurons (NeuN) co-localized with eGFP in cervical spinal cord sections of Chat-L10a-eGFP mice. Experiment was repeated two times with similar results. Scale bar, 50 µm. b, Number of motor neurons in cervical spinal cord sections of healthy (n = 3) and acute EAE (n = 4) Chat-L10a-eGFP mice with quantification. Student’s t-test, one-tailed: n = 3 healthy versus n = 4 EAE mice, t(5) = 2.440, P = 0.0293. Bars show mean values plus SEM. Scale bar, 100 µm. c, Principal component analysis including the top 500 variable genes. Experimental groups: spinal cord (SC), inflamed spinal cord (iSC), motor neurons (MN), inflamed motor neurons (iMN). d, Heatmap of Eeuclidean sample distances with unsupervised hierarchical clustering. Colors of experimental groups as in (c). *P < 0.05; **P < 0.01.
Supplementary Fig. 2 Expression changes in inflamed spinal cords.
a, Comparison of inflamed spinal cords (iSC) versus healthy spinal cords (SC). Bar graph of differentially expressed (DE) genes. Red, up-regulated, Blue down-regulated. Darker tones of red and blue represent increasing absolute log2 fold changes (>1, >2, >4). Candidates were identified by DESeq2, negative binomial generalized linear models, adjusted for multiple comparisons by FDR: n = 5 biologically independent samples per group each pooled from 3 mice, log2 fold change > 1, FDR-adjusted P < 0.05. b, Percentage of DE genes annotated for the gene ontology (GO) term ‘immune system process’. c, Gene expression heatmap of top 20 up- and down-regulated genes. Genes annotated for ‘immune system process’ are marked in ocher on the left. d, Enrichment map of regulated biological process GO terms. Red, up-regulated. Blue, down-regulated. Node size, gene set size. Grey lines, gene set overlap.
Supplementary Fig. 3 Expression changes in inflamed motor neurons and candidate gene identification.
a, Comparison of inflamed motor neurons (iMN) versus healthy motor neurons (MN). Bar graph of differentially expressed (DE) genes. Red, up-regulated, Blue down-regulated. Darker tones of red and blue represent increasing absolute log2 fold changes (>1, >2, >4). Candidates were identified by DESeq2, negative binomial generalized linear models, adjusted for multiple comparisons by FDR: n = 5 biologically independent samples per group each pooled from 3 mice, log2 fold change > 1, FDR-adjusted P < 0.05. b, Percentage of DE genes annotated for the gene ontology (GO) term ‘immune system process’. c, Gene expression heatmap of top 20 up- and down-regulated genes. Genes annotated for ‘immune system process’ are marked in ocher on the left. d, Enrichment map of regulated biological process GO terms. Red, up-regulated. Blue, down-regulated. Node size, gene set size. Grey lines, gene set overlap. e, Venn diagrams summarizing candidate criteria. f, Bar graph of candidate genes. Red, up-regulated, Blue down-regulated. Darker tones of red and blue represent increasing absolute log2 fold changes (>1, >2, >4). g, Percentage of candidate genes annotated for the GO term ‘immune system process’.
Supplementary Fig. 4 Candidate genes drive GO enrichment analysis.
a–b, Heatmap of candidate gene expression in healthy motor neurons (MN) and inflamed motor neurons (iMN) belonging to the regulated gene ontology (GO) terms ‘mitochondrion’ and ‘ubiquitin-dependent protein catabolic process’.
Supplementary Fig. 5 Characterization of Bsn expression.
a, Analysis of BSN expression in human MS microarray data from Han, M. H. et al. J Exp Med 2012. b, Gene expression analysis of Bsn in an independent EAE TRAP cohort (n = 3 per group). MN, motor neurons. iMN, inflamed motor neurons. Student’s t-test, two-tailed: n = 4 MN versus n = 3 iMN samples each pooled from 3 mice, t(5) = 2.695, P = 0.0431. Bars show mean values plus SEM. c, Immunohistochemical stainings of Bsn with quantification in spinal cord sections of healthy and chronic EAE mice at day 30 post immunization. Co-staining for neurons (NeuN). Student’s t-test, two-tailed: n = 3 mice per group, t(4) = 4.062, P = 0.0153. Bars show mean values plus SEM. MFI, mean fluorescence intensity. Scale bar, 20 µm. d, Immunohistochemical staining of Bsn in spinal cord sections of animals immunized with complete Freund’s adjuvant but without MOG35–55 at day 15 post immunization. Co-staining for neurons (NeuN). Experiment was repeated two times with similar results. Scale bar, 20 µm. e, Immunohistochemical staining of Bsn in spinal cord sections of EAE animals at day 15 post immunization. Co-staining for neurons (NeuN). Experiment was repeated three times with similar results. Scale bar, 20 µm. f, Immunohistochemical stainings of synapsin 1/2 in cervical spinal cord sections of healthy and chronic EAE mice. Co-staining for neurons (NeuN) and bassoon (Bsn). Experiment was repeated two times with similar results. Scale bar, 20 µm. *P < 0.05.
Supplementary Fig. 6 Staining of BSN in MS tissue.
a, Immunohistochemical stainings of exemplary MS lesions showing demyelination (loss of myelin basic protein, MBP) and varying degrees of inflammatory activity (CD3+ T cells, CD68+ activated phagocytes). Scale bar, 100 µm. b, Immunohistochemical staining of BSN in comparison to control stainings without BSN primary antibody in MS tissue. Co-staining for neurons (MAP2). Scale bar, 50 µm. c, Quantification of immunohistochemical stainings for BSN in spinal cord sections of control individuals and MS patients. 5–10 high power fields from 4 MS patients (n = 27 power fields) and 3 controls (n = 16 power fields). Median with interquartile range. d, Analysis of mouse Bsn and human BSN amino acid sequence for disordered residues by PONDR VSL2 algorithm. Red value indicates the percentage of disordered residues.
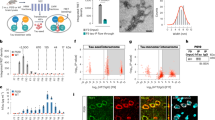
Supplementary Fig. 7 Overexpression of eGFP-Bsn in N2a cells.
a, Somatic distribution of eGFP signal in N2a cells overexpressing eGFP, eGFP–Bsn or eGFP–Map1a. Experiment was repeated three times with similar results. Scale bar, 20 µm. b, Co-localization of punctate eGFP–Bsn accumulation with markers for Golgi (Golgin97), endosomes (Rab5), ER (PDI) and lysosomes (LAMP-2). Scale bar, 20 µm. c, Overlap of genes regulated in EAE motor neurons (EAE candidates) and eGFP–Bsn overexpressing N2a cells (Bsn overexpression). Statistical analysis was performed by hypergeometric testing. d, Overlap of GO terms regulated in EAE motor neurons (EAE candidates) and eGFP–Bsn overexpressing N2a cells (Bsn overexpression). Statistical analysis was performed by hypergeometric testing.
Supplementary Fig. 8 Neuroaxonal numbers and immune cell infiltration in Bsn−/−and IU1 treatment EAE.
a, Quantification of neurofilament-positive axons in the dorsal columns of cervical spinal cord sections of wildtype and Bsn−/− animals without EAE induction. Student’s t-test, two-tailed: n = 3 mice per group, t(4) = 1.165, P = 0.3086. Bars show mean values plus SEM. b, Quantification of NeuN-positive neurons in cervical spinal cord sections of wildtype and Bsn−/− animals without EAE induction. Student’s t-test, two-tailed: n = 3 mice per group, t(4) = 0.01243, P = 0.9907. Bars show mean values plus SEM. c, Gating strategy of CNS-infiltrating T cells. d, T cell subset composition in CNS infiltrates of wildtype and Bsn−/− EAE animals at day 15 after immunization. Two-way ANOVA with Sidak’s post hoc test: n = 3 mice per group, F(2,12) = 0.0060, P = 0.9940; Sidak’s post hoc test: CD8+ T cells, WT versus Bsn−/−: P = 0.9998; CD4+ T cells, WT versus Bsn−/−: P = 0.9998; CD4+Foxp3+ Treg, WT versus Bsn−/−: P > 0.9999. Bars show mean values plus SEM. e, T cell subset composition in CNS infiltrates of vehicle and IU1-treated EAE animals at day 15 after immunization. Two-way ANOVA with Sidak’s post hoc test: n = 3 mice per group, F(2,12) = 6.666, P = 0.0113; Sidak’s post hoc test: CD8+ T cells, vehicle-treated versus IU1-treated: P = 0.0778; CD4+ T cells, vehicle-treated versus IU1-treated: P = 0.0778; CD4+Foxp3+ Treg, vehicle-treated versus IU1-treated: P = 0.7514.Bars show mean values plus SEM.
Supplementary Fig. 9 Graphical summary.
To investigate the neuronal response to central nervous system (CNS) inflammation, ribosome-bound mRNA from spinal cord motor neurons was isolated by translating ribosome affinity purification (TRAP) comparing healthy mice to mice undergoing experimental autoimmune encephalomyelitis (EAE), the animal model of multiple sclerosis (MS). Samples were subjected to RNA-seq and regulated candidate genes and associated pathways were identified (top). Results indicate a cytoplasmic deposition of the strongly induced presynaptic protein bassoon (Bsn) in both EAE and MS that was accompanied by induction of protein catabolism and reduced energy metabolism. Bsn overexpression and genetic deletion established a toxic gain-of-function of Bsn that drives neurodegeneration during CNS inflammation. Pharmacological activation of the proteasome cleared Bsn deposits and enhanced neuronal survival (bottom).
Supplementary information
Supplementary Table 1
Custom gene sets.
Supplementary Table 2
GSEA analysis.
Supplementary Table 3
Candidate lists.
Supplementary Table 4
Toppgene analysis.
Supplementary Table 5
MS–EAE overlap.
Supplementary Table 6
MS subjects.
Supplementary Table 7
Single cell analysis.
Rights and permissions
About this article
Cite this article
Schattling, B., Engler, J.B., Volkmann, C. et al. Bassoon proteinopathy drives neurodegeneration in multiple sclerosis. Nat Neurosci 22, 887–896 (2019). https://doi.org/10.1038/s41593-019-0385-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41593-019-0385-4
This article is cited by
-
hnRNP A1 dysfunction alters RNA splicing and drives neurodegeneration in multiple sclerosis (MS)
Nature Communications (2024)
-
Magnetic transferrin nanoparticles (MTNs) assay as a novel isolation approach for exosomal biomarkers in neurological diseases
Biomaterials Research (2023)
-
Bidirectional Communication Between the Brain and Other Organs: The Role of Extracellular Vesicles
Cellular and Molecular Neurobiology (2023)
-
Targeting the TCA cycle can ameliorate widespread axonal energy deficiency in neuroinflammatory lesions
Nature Metabolism (2023)
-
Mechanism-based criteria to improve therapeutic outcomes in progressive multiple sclerosis
Nature Reviews Neurology (2022)