Abstract
Memories of fearful events can last a lifetime. The prelimbic (PL) cortex, a subregion of prefrontal cortex, plays a critical role in fear memory retrieval over time. Most studies have focused on acquisition, consolidation, and retrieval of recent memories, but much less is known about the neural mechanisms of remote memory. Using a new knock-in mouse for activity-dependent genetic labeling (TRAP2), we demonstrate that neuronal ensembles in the PL cortex are dynamic. PL neurons TRAPed during later memory retrievals are more likely to be reactivated and make larger behavioral contributions to remote memory retrieval compared to those TRAPed during learning or early memory retrieval. PL activity during learning is required to initiate this time-dependent reorganization in PL ensembles underlying memory retrieval. Finally, while neurons TRAPed during earlier and later retrievals have similar broad projections throughout the brain, PL neurons TRAPed later have a stronger functional recruitment of cortical targets.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Code availability
Custom code for axon quantification, tSNE, and PCA analyses are available from the authors upon reasonable request.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Maren, S. Neurobiology of Pavlovian fear conditioning. Annu. Rev. Neurosci. 24, 897–931 (2001).
Frankland, P. W. & Bontempi, B. The organization of recent and remote memories. Nat. Rev. Neurosci. 6, 119–130 (2005).
Do Monte, F. H., Quirk, G. J., Li, B. & Penzo, M. A. Retrieving fear memories, as time goes by…. Mol. Psychiatry 21, 1027–1036 (2016).
Gale, G. D. et al. Role of the basolateral amygdala in the storage of fear memories across the adult lifetime of rats. J. Neurosci. 24, 3810–3815 (2004).
Grewe, B. F. et al. Neural ensemble dynamics underlying a long-term associative memory. Nature 543, 670–675 (2017).
Todd, T. P., Mehlman, M. L., Keene, C. S., DeAngeli, N. E. & Bucci, D. J. Retrosplenial cortex is required for the retrieval of remote memory for auditory cues. Learn. Mem. 23, 278–288 (2016).
Do-Monte, F. H., Quiñones-Laracuente, K. & Quirk, G. J. A temporal shift in the circuits mediating retrieval of fear memory. Nature 519, 460–463 (2015).
Sierra-Mercado, D., Padilla-Coreano, N. & Quirk, G. J. Dissociable roles of prelimbic and infralimbic cortices, ventral hippocampus, and basolateral amygdala in the expression and extinction of conditioned fear. Neuropsychopharmacology 36, 529–538 (2011).
Corcoran, K. A. & Quirk, G. J. Activity in prelimbic cortex is necessary for the expression of learned, but not innate, fears. J. Neurosci. 27, 840–844 (2007).
Courtin, J. et al. Prefrontal parvalbumin interneurons shape neuronal activity to drive fear expression. Nature 505, 92–96 (2014).
Kitamura, T. et al. Engrams and circuits crucial for systems consolidation of a memory. Science 356, 73–78 (2017).
Lesburguères, E. et al. Early tagging of cortical networks is required for the formation of enduring associative memory. Science 331, 924–928 (2011).
Tse, D. et al. Schema-dependent gene activation and memory encoding in neocortex. Science 333, 891–895 (2011).
Maviel, T., Durkin, T. P., Menzaghi, F. & Bontempi, B. Sites of neocortical reorganization critical for remote spatial memory. Science 305, 96–99 (2004).
Wheeler, A. L. et al. Identification of a functional connectome for long-term fear memory in mice. PLoS Comput. Biol. 9, e1002853 (2013).
DeNardo, L. & Luo, L. Genetic strategies to access activated neurons. Curr. Opin. Neurobiol. 45, 121–129 (2017).
Vertes, R. P. Differential projections of the infralimbic and prelimbic cortex in the rat. Synapse 51, 32–58 (2004).
Gabbott, P. L. A., Warner, T. A., Jays, P. R. L., Salway, P. & Busby, S. J. Prefrontal cortex in the rat: projections to subcortical autonomic, motor, and limbic centers. J. Comp. Neurol. 492, 145–177 (2005).
Karalis, N. et al. 4-Hz oscillations synchronize prefrontal-amygdala circuits during fear behavior. Nat. Neurosci. 19, 605–612 (2016).
Senn, V. et al. Long-range connectivity defines behavioral specificity of amygdala neurons. Neuron 81, 428–437 (2014).
Sotres-Bayon, F., Sierra-Mercado, D., Pardilla-Delgado, E. & Quirk, G. J. Gating of fear in prelimbic cortex by hippocampal and amygdala inputs. Neuron 76, 804–812 (2012).
Popa, D., Duvarci, S., Popescu, A. T., Léna, C. & Paré, D. Coherent amygdalocortical theta promotes fear memory consolidation during paradoxical sleep. Proc. Natl. Acad. Sci. USA. 107, 6516–6519 (2010).
Guenthner, C. J., Miyamichi, K., Yang, H. H., Heller, H. C. & Luo, L. Permanent genetic access to transiently active neurons via TRAP: targeted recombination in active populations. Neuron 78, 773–784 (2013).
Allen, W. E. et al. Thirst-associated preoptic neurons encode an aversive motivational drive. Science 357, 1149–1155 (2017).
Coulon, V., Chebli, K., Cavelier, P. & Blanchard, J. M. A novel mouse c-fos intronic promoter that responds to CREB and AP-1 is developmentally regulated in vivo. PLoS One 5, e11235 (2010).
Wilson, T. & Treisman, R. Removal of poly(A) and consequent degradation of c-fos mRNA facilitated by 3′ AU-rich sequences. Nature 336, 396–399 (1988).
Shimshek, D. R. et al. Codon-improved Cre recombinase (iCre) expression in the mouse. Genesis 32, 19–26 (2002).
Madisen, L. et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 (2010).
Hess, U. S., Lynch, G. & Gall, C. M. Regional patterns of c-fos mRNA expression in rat hippocampus following exploration of a novel environment versus performance of a well-learned discrimination. J. Neurosci. 15, 7796–7809 (1995).
Renier, N. et al. Mapping of brain activity by automated volume analysis of immediate early genes. Cell 165, 1789–1802 (2016).
Tovote, P., Fadok, J. P. & Lüthi, A. Neuronal circuits for fear and anxiety. Nat. Rev. Neurosci. 16, 317–331 (2015).
LeDoux, J. E. Emotion circuits in the brain. Annu. Rev. Neurosci. 23, 155–184 (2000).
Anagnostaras, S. G., Maren, S. & Fanselow, M. S. Temporally graded retrograde amnesia of contextual fear after hippocampal damage in rats: within-subjects examination. J. Neurosci. 19, 1106–1114 (1999).
Lee, J. H. et al. Global and local fMRI signals driven by neurons defined optogenetically by type and wiring. Nature 465, 788–792 (2010).
Maren, S., Phan, K. L. & Liberzon, I. The contextual brain: implications for fear conditioning, extinction and psychopathology. Nat. Rev. Neurosci. 14, 417–428 (2013).
Berndt, A. et al. Structural foundations of optogenetics: determinants of channelrhodopsin ion selectivity. Proc. Natl Acad. Sci. USA. 113, 822–829 (2016).
Bero, A. W. et al. Early remodeling of the neocortex upon episodic memory encoding. Proc. Natl Acad. Sci. USA. 111, 11852–11857 (2014).
Armbruster, B., Li, X., Pausch, M. H., Herlitze, S. & Roth, B. L. Evolving the lock to fit the key to create a family of G protein-coupled receptors potently activated by an inert ligand. Proc. Natl Acad. Sci. USA. 104, 5163–5168 (2007).
van der Maaten, L. & Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008).
Frankland, P. W., Bontempi, B., Talton, L. E., Kaczmarek, L. & Silva, A. J. The involvement of the anterior cingulate cortex in remote contextual fear memory. Science 304, 881–883 (2004).
Sacco, T. & Sacchetti, B. Role of secondary sensory cortices in emotional memory storage and retrieval in rats. Science 329, 649–656 (2010).
Takehara-Nishiuchi, K. Entorhinal cortex and consolidated memory. Neurosci. Res. 84, 27–33 (2014).
Hoover, W. B. & Vertes, R. P. Anatomical analysis of afferent projections to the medial prefrontal cortex in the rat. Brain. Struct. Funct. 212, 149–179 (2007).
DeNardo, L. A., Berns, D. S., DeLoach, K. & Luo, L. Connectivity of mouse somatosensory and prefrontal cortex examined with trans-synaptic tracing. Nat. Neurosci. 18, 1687–1697 (2015).
Ziv, Y. et al. Long-term dynamics of CA1 hippocampal place codes. Nat. Neurosci. 16, 264–266 (2013).
Driscoll, L. N., Pettit, N. L., Minderer, M., Chettih, S. N. & Harvey, C. D. Dynamic reorganization of neuronal activity patterns in parietal cortex. Cell 170, 986–999.e16 (2017).
Burgos-Robles, A., Vidal-Gonzalez, I. & Quirk, G. J. Sustained conditioned responses in prelimbic prefrontal neurons are correlated with fear expression and extinction failure. J. Neurosci. 29, 8474–8482 (2009).
Vetere, G. et al. Chemogenetic interrogation of a brain-wide fear memory network in mice. Neuron 94, 363–374.e4 (2017).
Preston, A. R. & Eichenbaum, H. Interplay of hippocampus and prefrontal cortex in memory. Curr. Biol. 23, R764–R773 (2013).
Paxinos, G. & Franklin, K. B. J. The Mouse Brain in Stereotaxic Coordinates. 4th edn, (Academic Press, Cambridge, MA, USA, 2001).
Beier, K. T. et al. Circuit Architecture of VTA dopamine neurons revealed by systematic input-output mapping. Cell 162, 622–634 (2015).
Zhou, T. et al. History of winning remodels thalamo-PFC circuit to reinforce social dominance. Science 357, 162–168 (2017).
Wayman, W. N. & Woodward, J. J. Chemogenetic excitation of accumbens-projecting infralimbic cortical neurons blocks toluene-induced conditioned place preference. J. Neurosci. 38, 1462–1471 (2018).
Shekhar, K. et al. Comprehensive classification of retinal bipolar neurons by single-cell transcriptomics. Cell 166, 1308–1323.e30 (2016).
Acknowledgements
We thank C. Malanac for mouse genotyping, the Stanford Transgenic Facility for help in producing TRAP2 knock-in mice and Stanford Viral Core, K. Deisseroth and C. Ramakrishnan for AAV vectors, T. Bonhoeffer, P. Goltstein, and members of the Luo Lab for comments on the manuscript. L.A.D. was supported by an NIH postdoctoral fellowship (F32NS087860) and a Mentored Research Scientist Development Award (K01MH11626401). W.E.A. was supported by a Fannie & John Hertz Foundation Fellowship and a National Science Foundation Graduate Research Fellowship (grant DGE-114747). E.L.A. was supported by the Standford Bio-X Ph.D Fellowship program and the William K. Bowles Jr. Foundation. L.L. is an HHMI investigator. This work was supported by grants from National Science Foundation, National Institutes of Health, and a Hughes Collaborative Innovation Award.
Author information
Authors and Affiliations
Contributions
L.A.D. and L.L. designed experiments. L.A.D. and C.J.G. generated the TRAP2 targeting construct. L.A.D. characterized the TRAP2 mouse. L.A.D. and C.D.L. performed behavior assays. L.A.D. and L.F. performed histology and confocal imaging. L.A.D., C.D.L., and L.F. analyzed the data, W.E.A. analyzed iDISCO+-generated data sets. E.L.A. (with support from M.T.-L.) advised and provided training in the iDISCO+ and ClearMap methods. D.F. wrote software and advised for quantitative whole-brain axon analysis. L.A.D. and L.L. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1
Targeting strategy for generating TRAP2 mice.
Supplementary Figure 2 Time course of TRAPing.
a, Timeline of visual stimulation experiment to determine effective TRAPing window. b, Example images of TRAPed cells in primary visual cortex (V1) and primary somatosensory cortex (S1) of TRAP2 mice that underwent the visual stimulation experiment. Scale bars, 100 µm. c, Quantification of TRAPed cell density in TRAP2 mice (V1, F(4,16) = 10.07, P = 0.0003, n = 4, 4, 4, 5, 4 for dark, –6h, –3h, 0h, and 3h conditions, respectively); S1, F(3,6) = 1.786, P = 0.249, n = 4, 3, 3, 4, 3 for dark, –6h, –3h, 0h, and 3h conditions, respectively). d, Quantification of fold change in TRAPed cells in TRAP2 mice (V1, F(4,17) = 10.85, P = 0.0001, n = 4, 4, 4, 5, 4 for dark, –6h, –3h, 0h, and 3h conditions, respectively; S1, F(4,12) = 2.590, P = 0.0903 n = 4, 3, 3, 4, 3 for dark, –6h, –3h, 0h, and 3h conditions, respectively; for both c and d, one-way ANOVA with Dunnett post-hoc test). e–g, Analogous experiments and analyses as b–d, but using TRAP1 mice. Statistics for f, V1, F(3,6) = 12.93, P = 0.0005; S1, F(3,6) = 1.786, P = 0.2497; , = 3 per condition. For g, V1, F(3,7) = 11.16, P = 0.0047; S1, F(3,6) = 1.786, P = 0.2497, n = 3 per condition. In all plots and statistical tests, n represents biologically independent animals. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 and summary graphs show mean±SEM. Scale bars, 100 µm.
Supplementary Figure 3 Sensitivity of TRAP2.
a, Schematic for novel environment (NE) experiment to determine sensitivity of TRAP2. b, Example coronal hemisections from TRAP2 (left), TRAP1 (middle), and TRAP2 mice injected with solvent instead of 4-OHT (right) from NE condition. Numbers beside the atlas diagrams represent anterior-posterior positions with respect to bregma. c, Quantification of TRAPed cells in motor cortex (MOp), primary somatosensory cortex (SSp), dorsal striatum (STRd), nucleus accumbens (ACB), lateral septum (LS), bed nucleus of the stria terminalis (BST), lateral hypothalamic area (LHA), basolateral amygdala (BLA), hippocampal field CA1, and periaqueductal grey (PAG) (F(2,58) = 101.7, P < 0.0001, n = 3 per condition, 2-way ANOVA with Dunnett post-hoc test). d, Quantification of TRAPed cell density in TRAP2 home-cage compared to TRAP1 home-cage conditions (F(1,40) = 13.33, P = 0.0007, n = 3 per condition, 2-way ANOVA with Sidak post-hoc test). Scale bars, 500 µm. In all plots and statistical tests, n represents biologically independent animals. Summary graphs show mean±SEM. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Supplementary Figure 4 Summary of behavior and TRAP/Fos activation in PL.
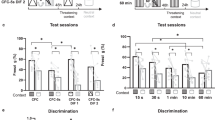
a, Summary of freezing behavior for all time points used throughout the study [FC, fear conditioning; Ret1, retrieval 1 (1d, 7d or 14d), Ret2, retrieval 2 (28d)]; Pre: before conditioning, CS+: conditioned tone; CS–: unreinforced tone; ctx: context). (FC: F = 61.31, P < 0.0001, Ret 1: F = 32.23, P < 0.0001, Ret 2: F = 37.3, P < 0.0001, One-way ANOVA with Tukey post-hoc test, n = 29). b, Quantification of freezing to CS+ for FC (n = 8), 1d (n = 8), 7d (n = 8), 14d (n = 4), and NS groups (n = 15). (FC: F = 69.58, P < 0.0001, Ret1: F = 36.66, P < 0.0001, Ret2: F = 7, P < 0.0001, One-way ANOVA with Tukey post-hoc test). c, Quantification of % TRAPed cells by group (F = 7.06, P = 0.0002, One-way ANOVA with Tukey post-hoc test). d, Quantification of % Fos+ cells by group (F = 1.59, P = 0.196, One-way ANOVA with Tukey post-hoc test). e, Comparison of Fos expression normalized to chance levels in TRAPed (colored bars, (Double+/DAPI)/((Fos+/DAPI)x(TRAP+/DAPI))) and non-TRAPed (black bars, (Double+/DAPI)/((Fos+/DAPI)x(TRAP–/DAPI))) PL neurons; stars represents results of multiple t-tests with Holm-sidak posthoc correction for multiple comparisons). For c–e, n = 15, 8, 8, 8, 8 for NS, FC, 1d, 7d, and 14d conditions, respectively. In all plots and statistical tests, n represents biologically independent animals. In b, boxes represent 25–75th percentile, whiskers represent 10–90th percentile, lines represent median. All other summary graphs show mean±SEM. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
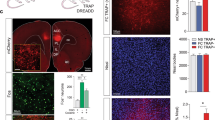
Supplementary Figure 5 Comparing Fos expression following remote fear memory retrieval in WT and TRAP2 animals.
a,d,g, Schematics showing locations of confocal imaging (blue). Example confocal stacks and summary graphs for b, prelimbic (PL) cortex [P = 0.36; WT, n = 3, TRAP2, n = 5], c, piriform cortex [P = 0.82; WT, n = 3, TRAP2, n = 6], e, paraventricular thalamus (PVT) [P = 0.82; WT, n = 3, TRAP2, n = 4], f, paraventricular hypothalamus (PVH) [P = 0.79, WT, n = 3, TRAP2 n = 3], h, basolateral amygdala (BLA) [P = 0.13, WT, n = 3, TRAP2, n = 10], i, dentate gyrus (DG) [P = 0.38; WT, n = 3, TRAP2, n = 7], two-sided unpaired t-tests. Scale bars, 100 µm. In all plots and statistical tests, n represents biologically independent animals. Summary graphs show mean±SEM. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Supplementary Figure 6 Analysis of activated neurons in PL by cortical layer.
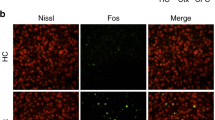
a, Example images of TRAPed and Fos+ neurons in PL from each experimental group. Insets show high-magnifications of example reactivated cells. Scale bars, 100 µm. b, Quantification of cortical layer thickness in each experimental condition [FGroup(3,24) = 0.777, P = 0.518, 2-way ANOVA with Tukey post-hoc test]. c, Quantification of active neurons in PL cortical layers in each experimental group, expressed as a fraction of total active neurons in PL per brain analyzed [FGroup(3,32) = 50.73, P < 0.0001, 2-way repeated measures ANOVA with Tukey post-hoc test]. d, Quantification of TRAPed neurons in PL cortical layers that are reactivated during 28d memory retrieval. Reactivated (Double+/TRAPed) cells per layer presented as a fraction of total reactivated cells counted in PL for each brain [FGroup(3,28) = 1.3, P = 0.294, 2-way ANOVA with Tukey post-hoc test]. For b–d, n = 14, 7, 7, 4, 4, 18 for FC, 1d, 7d, 14d, and 28d (Fos) conditions, respectively. In all plots and statistical tests, n represents biologically independent animals. Summary graphs show mean±SEM. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Supplementary Figure 7 TRAP/Fos activation in piriform cortex.
a, Example confocal images of TRAPed and Fos+ neurons in piriform cortex from each experimental group as in Fig. 2. Scale bars, 100 µm. b, Quantification of TRAPed cells per section (F = = 6.588, P = 0.0149). c, Quantification of Fos+ cells / section (F = 0.3839, P = 0.7676). d, Quantification of Double+ / Fos+ cells (F = 2.772, P = 0.1106). e, Quantification of Double+/TRAPed cells per section (F = 0.3604, P = 0.7834). One-way ANOVA with Tukey post-hoc test, n = 3 for all conditions. In all plots and statistical tests, n represents biologically independent animals. Summary graphs show mean±SEM. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Supplementary Figure 8 Additional optogenetics and behavior analyses.
a, Experimental protocol for fear conditioning and testing the photoactivation effect of NS-TRAPed animals. b,c, Quantification of contextual (b, P = 0.103, n = 9, two-sided paired t-test) and CS+-evoked (c, P = 0.938, n = 9, two-sided paired t-test) freezing in NS animals that were subsequently fear conditioned in response to photoactivation of NS-TRAPed cells. d–h, Freezing levels with or without (±) ChR2 activation for individual animals summarized in Fig. 3. d, freezing to CS+ (FC, P = 0.012, n = 18; 1d, P = 0.410, n = 19; 7d, P = 0.182, n = 10; 14d, P = 0.250, n = 10; two-sided paired t-tests. e, freezing to CS– (FC, P = 0.731, n = 11; 1d, P = 0.710, n = 13; 7d, P = 0.078, n = 10; 14d, P = 0.947, n = 10; two-sided paired t-tests). f, tone discrimination index (FC, P = 0.597, n = 11; 1d, P = 0.478, n = 13; 7d, P = 0.233, n = 10; 14d, P = 0.858, n = 10; two-sided paired t-tests). g, freezing to altered context (FC, P = 0.093, n = 11; 1d, P = 0.039, n = 13; 7d, P = 0.021, n = 12; 14d, P = 0.100, n = 8; two-sided paired t-tests). h, freezing to CS+ in altered context (FC, P = 0.466, n = 11; 1d, P = 0.268, n = 13; 7d, P = 0.705, n = 11; 14d, P = 0.039, n = 8; two-sided paired t-tests). In all plots and statistical tests, n represents biologically independent animals. Summary graphs show mean±SEM. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Supplementary Figure 9 Histological analysis of fiber placement in PL in optogenetic activation experiments.
a, Representative slide scanner images of PL showing optical fiber placement and ChR2-eYFP injections across experimental groups. b, Cre-dependent virus did not express in solvent-injected animals (no 4-OHT). c, Mapping ChR2 optical fiber locations to a standard mouse brain atlas (Paxinos, G. & Franklin, K.B.J. The Mouse Brain in Stereotaxic Coordinates, 2nd ed. Acadmic Press, 2001). Each circle represents the position of optic fiber termination site of one mouse. Colors correspond to TRAPing group. Scale bars, 500 µm.
Supplementary Figure 10 No effect of stimulating TRAPed PL neurons in real-time place-aversion assay.
a, Representative animal tracks from five TRAP2 groups during 15 min stimulation session (Day 2). b, Quantification of aversive behavior [% time on unstimulated side during photostimulation session (Day 2) – % time on unstimulated side during baseline session (Day 1)] (F(4,51) = 1.702, P = 0.1639, One-way ANOVA with post-hoc Tukey test, n = 9, 8, 17, 12, 9 for NS, FC, 1d, 7d, 14, respectively). n represents biologically independent animals. Summary graph shows mean±SEM. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Supplementary Figure 11 Histological analysis of fiber placements in opto- and chemogenetic inhibition experiments.
a, Example image of iC++ expression and dual fiber implant in 1d-TRAPed brain. b, Fiber placement for 1d- and 14d-TRAPed iC++ brains. c, Fiber placement for hM4D experiments.
Supplementary Figure 12 Correlations between whole-brain TRAPing patterns and behavior.
Pearson correlations of regional TRAP counts with context freezing (a,b) or CS+-evoked freezing (c,d) color mapped onto tSNE clusters (as shown in Fig. 5c), or onto coronal sections from a standard brain for 1d-TRPAed (a,c) and 14d-TRAPed (b,d) brains (1d, n = 7; 14d, n = 9 biologically independent animals, see also Supplementary Table 2).
Supplementary information
Supplementary text and figures
Supplementary Figs. 1–12 and Supplementary Tables 1–4
Rights and permissions
About this article
Cite this article
DeNardo, L.A., Liu, C.D., Allen, W.E. et al. Temporal evolution of cortical ensembles promoting remote memory retrieval. Nat Neurosci 22, 460–469 (2019). https://doi.org/10.1038/s41593-018-0318-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41593-018-0318-7
This article is cited by
-
Dynamic and selective engrams emerge with memory consolidation
Nature Neuroscience (2024)
-
Context memory formed in medial prefrontal cortex during infancy enhances learning in adulthood
Nature Communications (2024)
-
Spatial transcriptomics reveal neuron–astrocyte synergy in long-term memory
Nature (2024)
-
Reactivation of encoding ensembles in the prelimbic cortex supports temporal associations
Neuropsychopharmacology (2024)
-
Dorsal peduncular cortex activity modulates affective behavior and fear extinction in mice
Neuropsychopharmacology (2024)