Abstract
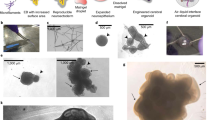
Investigating human oligodendrogenesis and the interaction of oligodendrocytes with neurons and astrocytes would accelerate our understanding of the mechanisms underlying white matter disorders. However, this is challenging because of the limited accessibility of functional human brain tissue. Here, we developed a new differentiation method of human induced pluripotent stem cells to generate three-dimensional brain organoids that contain oligodendrocytes as well as neurons and astrocytes, called human oligodendrocyte spheroids. We found that oligodendrocyte lineage cells derived in human oligodendrocyte spheroids transitioned through developmental stages similar to primary human oligodendrocytes and that the migration of oligodendrocyte lineage cells and their susceptibility to lysolecithin exposure could be captured by live imaging. Moreover, their morphology changed as they matured over time in vitro and started myelinating neurons. We anticipate that this method can be used to study oligodendrocyte development, myelination, and interactions with other major cell types in the CNS.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Gene expression data have been deposited in the Gene Expression Omnibus under accession number GSE115011. The data that support the findings of this study are available on reasonable request from the corresponding author.
References
Nave, K. A. Myelination and the trophic support of long axons. Nat. Rev. Neurosci. 11, 275–283 (2010).
Menichella, D. M. et al. Genetic and physiological evidence that oligodendrocyte gap junctions contribute to spatial buffering of potassium released during neuronal activity. J. Neurosci. 26, 10984–10991 (2006).
Fünfschilling, U. et al. Glycolytic oligodendrocytes maintain myelin and long-term axonal integrity. Nature 485, 517–521 (2012).
Dai, X. et al. The trophic role of oligodendrocytes in the basal forebrain. J. Neurosci. 23, 5846–5853 (2003).
Orthmann-Murphy, J. L., Freidin, M., Fischer, E., Scherer, S. S. & Abrams, C. K. Two distinct heterotypic channels mediate gap junction coupling between astrocyte and oligodendrocyte connexins. J. Neurosci. 27, 13949–13957 (2007).
Moore, C. S., Abdullah, S. L., Brown, A., Arulpragasam, A. & Crocker, S. J. How factors secreted from astrocytes impact myelin repair. J. Neurosci. Res. 89, 13–21 (2011).
Simons, M. & Trajkovic, K. Neuron-glia communication in the control of oligodendrocyte function and myelin biogenesis. J. Cell Sci. 119, 4381–4389 (2006).
Barres, B. A., Schmid, R., Sendnter, M. & Raff, M. C. Multiple extracellular signals are required for long-term oligodendrocyte survival. Development 118, 283–295 (1993).
Lin, S. C. & Bergles, D. E. Synaptic signaling between GABAergic interneurons and oligodendrocyte precursor cells in the hippocampus. Nat. Neurosci. 7, 24–32 (2004).
Bergles, D. E., Roberts, J. D., Somogyi, P. & Jahr, C. E. Glutamatergic synapses on oligodendrocyte precursor cells in the hippocampus. Nature 405, 187–191 (2000).
Hardy, R. & Reynolds, R. Neuron-oligodendroglial interactions during central nervous system development. J. Neurosci. Res. 36, 121–126 (1993).
van der Knaap, M. S., Pronk, J. C. & Scheper, G. C. Vanishing white matter disease. Lancet Neurol. 5, 413–423 (2006).
Wolswijk, G. Oligodendrocyte survival, loss and birth in lesions of chronic-stage multiple sclerosis. Brain 123, 105–115 (2000).
Douvaras, P. & Fossati, V. Generation and isolation of oligodendrocyte progenitor cells from human pluripotent stem cells. Nat. Protoc. 10, 1143–1154 (2015).
Numasawa-Kuroiwa, Y. et al. Involvement of ER stress in dysmyelination of Pelizaeus–Merzbacher disease with PLP1 missense mutations shown by iPSC-derived oligodendrocytes. Stem Cell Rep. 2, 648–661 (2014).
Stacpoole, S. R. et al. High yields of oligodendrocyte lineage cells from human embryonic stem cells at physiological oxygen tensions for evaluation of translational biology. Stem Cell Rep. 1, 437–450 (2013).
Wang, S. et al. Human iPSC-derived oligodendrocyte progenitor cells can myelinate and rescue a mouse model of congenital hypomyelination. Cell Stem Cell 12, 252–264 (2013).
Hogberg, H. T. et al. Toward a 3D model of human brain development for studying gene/environment interactions. Stem Cell Res. Ther. 4(Suppl 1), S4 (2013).
Pașca, S. P. The rise of three-dimensional human brain cultures. Nature 553, 437–445 (2018).
Paşca, A. M. et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods 12, 671–678 (2015).
Birey, F. et al. Assembly of functionally integrated human forebrain spheroids. Nature 545, 54–59 (2017).
Sloan, S. A., Andersen, J., Pașca, A. M., Birey, F. & Pașca, S. P. Generation and assembly of human brain region-specific three-dimensional cultures. Nat. Protoc. 13, 2062–2085 (2018).
Darmanis, S. et al. A survey of human brain transcriptome diversity at the single cell level. Proc. Natl Acad. Sci. USA 112, 7285–7290 (2015).
Sloan, S. A. et al. Human astrocyte maturation captured in 3D cerebral cortical spheroids derived from pluripotent stem cells. Neuron 95, 779–790.e6 (2017).
van der Maaten, L. & Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008).
Trapnell, C. et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014).
Zhang, Y. et al. Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron 89, 37–53 (2016).
Crow, Y. J. et al. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi–Goutières syndrome and mimic congenital viral brain infection. Nat. Genet. 38, 910–916 (2006).
Gomez-Ospina, N. Arylsulfatase A deficiency. in GeneReviews (eds. Adam, M. P. et al.) (Univ. Washington, Seattle, 2006).
Austin, J. et al. Studies in globoid (Krabbe) leukodystrophy (GLD). V. Controlled enzymic studies in ten human cases. Arch. Neurol. 23, 502–512 (1970).
Bergles, D. E. & Richardson, W. D. Oligodendrocyte development and plasticity. Cold Spring Harb. Perspect. Biol. 8, a020453 (2015).
Pol, S. U. et al. Sox10-MCS5 enhancer dynamically tracks human oligodendrocyte progenitor fate. Exp. Neurol. 247, 694–702 (2013).
Sontheimer, H., Trotter, J., Schachner, M. & Kettenmann, H. Channel expression correlates with differentiation stage during the development of oligodendrocytes from their precursor cells in culture. Neuron 2, 1135–1145 (1989).
Livesey, M. R. et al. Maturation and electrophysiological properties of human pluripotent stem cell-derived oligodendrocytes. Stem Cells 34, 1040–1053 (2016).
Káradóttir, R. & Attwell, D. Neurotransmitter receptors in the life and death of oligodendrocytes. Neuroscience 145, 1426–1438 (2007).
Káradóttir, R., Cavelier, P., Bergersen, L. H. & Attwell, D. NMDA receptors are expressed in oligodendrocytes and activated in ischaemia. Nature 438, 1162–1166 (2005).
Birgbauer, E., Rao, T. S. & Webb, M. Lysolecithin induces demyelination in vitro in a cerebellar slice culture system. J. Neurosci. Res. 78, 157–166 (2004).
Blakemore, W. F., Eames, R. A., Smith, K. J. & McDonald, W. I. Remyelination in the spinal cord of the cat following intraspinal injections of lysolecithin. J. Neurol. Sci. 33, 31–43 (1977).
Hall, S. M. The effect of injections of lysophosphatidyl choline into white matter of the adult mouse spinal cord. J. Cell Sci. 10, 535–546 (1972).
Amin, N. D. & Paşca, S. P. Building models of brain disorders with three-dimensional organoids. Neuron 100, 389–405 (2018).
Di Lullo, E. & Kriegstein, A. R. The use of brain organoids to investigate neural development and disease. Nat. Rev. Neurosci. 18, 573–584 (2017).
Yoon, S. et al. Reliability of human cortical organoid generation. Nat. Methods 16, 75–78 (2019).
Volpe, J. J. The encephalopathy of prematurity – brain injury and impaired brain development inextricably intertwined. Semin. Pediatr. Neurol. 16, 167–178 (2009).
Paşca, S. P. et al. Using iPSC-derived neurons to uncover cellular phenotypes associated with Timothy syndrome. Nat. Med. 17, 1657–1662 (2011).
Yazawa, M. et al. Using induced pluripotent stem cells to investigate cardiac phenotypes in Timothy syndrome. Nature 471, 230–234 (2011).
Gallego Romero, I. et al. A panel of induced pluripotent stem cells from chimpanzees: a resource for comparative functional genomics. eLife 4, e07103 (2015).
Marchetto, M. C. et al. Non-cell-autonomous effect of human SOD1 G37R astrocytes on motor neurons derived from human embryonic stem cells. Cell Stem Cell 3, 649–657 (2008).
Xue, Y. et al. Sequential regulatory loops as key gatekeepers for neuronal reprogramming in human cells. Nat. Neurosci. 19, 807–815 (2016).
The HD iPSC Consortium. Developmental alterations in Huntington’s disease neural cells and pharmacological rescue in cells and mice. Nat. Neurosci. 20, 648–660 (2017).
Renner, M. et al. Self-organized developmental patterning and differentiation in cerebral organoids. EMBO J. 36, 1316–1329 (2017).
Kang, S. M. et al. Efficient induction of oligodendrocytes from human embryonic stem cells. Stem Cells 25, 419–424 (2007).
Deborde, S. et al. Schwann cells induce cancer cell dispersion and invasion. J. Clin. Invest. 126, 1538–1554 (2016).
Xiao, D. et al. Direct reprogramming of fibroblasts into neural stem cells by single non-neural progenitor transcription factor Ptf1a. Nat. Commun. 9, 2865 (2018).
Clarke, K. E. et al. A robust and reproducible human pluripotent stem cell derived model of neurite outgrowth in a three-dimensional culture system and its application to study neurite inhibition. Neurochem. Int. 106, 74–84 (2017).
Greber, B. et al. FGF signalling inhibits neural induction in human embryonic stem cells. EMBO J. 30, 4874–4884 (2011).
Amin, H. et al. Electrical responses and spontaneous activity of human iPS-derived neuronal networks characterized for 3-month culture with 4096-electrode arrays. Front. Neurosci. 10, 121 (2016).
Picelli, S. et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014).
Risso, D., Ngai, J., Speed, T. P. & Dudoit, S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat. Biotechnol. 32, 896–902 (2014).
Penna, I. et al. Selection of candidate housekeeping genes for normalization in human postmortem brain samples. Int. J. Mol. Sci. 12, 5461–5470 (2011).
Fan, J. et al. Characterizing transcriptional heterogeneity through pathway and gene set overdispersion analysis. Nat. Methods 13, 241–244 (2016).
Deverman, B. E. et al. Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat. Biotechnol. 34, 204–209 (2016).
Thomas, C. A. et al. Modeling of TREX1-dependent autoimmune disease using human stem cells highlights L1 accumulation as a source of neuroinflammation. Cell Stem Cell 21, 319–331.e8 (2017).
Mariani, J. et al. FOXG1-dependent dysregulation of GABA/glutamate neuron differentiation in autism spectrum disorders. Cell 162, 375–390 (2015).
Acknowledgements
This paper is dedicated to the memory of our wonderful colleague and mentor Ben A. Barres. We thank B.A. Barres, B. Zuchero, and members of the Pasca laboratory for scientific input, J. Perrino (Stanford Cell Sciences Imaging Facility) for support with electron microscopy, as well as F. Sim (University of Buffalo) for providing the Sox10-MCS5::eGFP plasmid. This work was supported by the US National Institutes of Health BRAINS Award (MH107800), the MQ Fellow Award, the NYSCF Robertson Stem Cell Investigator Award, the Stanford Wu Tsai Neurosciences Institute’s Brain Rejuvenation Project and the Human Brain Organogenesis Project, the Kwan Research Fund and the California Institute of Regenerative Medicine, the Child Health Research Institute Pilot Award, and the NARSAD Independent Investigator Award from the Brain and Behavior Research Foundation (to S.P.P.); the National Science Foundation Graduate Research Fellowship and the Bio-X Stanford Interdisciplinary Graduate Fellowship (to R.M.M.); Stanford Medicine’s Dean’s Fellowship (to Y.M.); and NIMH T32GM007365, F30MH106261, and Bio-X Predoctoral Fellowship (to or supporting S.A.S.).
Author information
Authors and Affiliations
Contributions
R.M.M. performed the differentiation experiments. Q.L. carried out single-cell library preparations and S.A.S. analyzed single-cell data. O.R. and J.R.H. conducted and analyzed electrophysiological experiments. R.M.M., Y.M., and R.J.L. carried out all other experiments and data analyses. R.M.M. and S.P.P. conceived the project, designed experiments, and wrote the manuscript with input from all authors. S.P.P. supervised the work.
Corresponding author
Ethics declarations
Competing interests
Stanford University has filed a provisional patent application that covers the generation of myelinating oligospheroids for studying human development and disease (US patent application number 15/953,197).
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Characterization of oligodendrocytes, astrocytes, and neurons in hOLS.
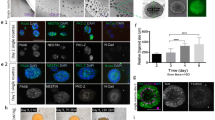
a, Relative gene expression (normalized to GAPDH) as determined by qPCR at day 25 of in vitro culture of markers for pluripotency and germ layers (OCT4, SOX2, BRACH, SOX17) (n = 4 RNA samples of 2 hOLS each from 4 hiPS cell lines; see Supplementary Table 1; Kruskal–Wallis test, P = 0.001). b, Relative gene expression (normalized to GAPDH) in hCS and hOLS as determined by qPCR at day 37 of in vitro culture hOLS of CNS markers (FOXG1, SIX3, NKX2.1, OTX2, LMX1B, RAX1, HOXB4, PAX6, and LHX2) (n = 5 samples of 2 hOLS each from 5 hiPS cell lines; see Supplementary Table 1; Kruskal–Wallis test, P = 0.0001). c, Day 51 immunostaining of hOLS cryosections for OLIG2 and NKX2-2. Immunostainings were repeated on hOLS from four independent inductions with similar results. d, Histogram of the spatial distribution of MBP+ cells in hOLS cryosections. The number of MBP+ cells at various proportions of the radius (distance from center to edge) is shown. e, Relative gene expression (normalized to GAPDH) as determined by qPCR at day 100 of in vitro culture in hCS and hOLS of RBFOX3 (NEUN) (two-tailed t-test, t = 1.23, d.f. = 15, P = 0.23) and GFAP (two-tailed Mann–Whitney test, **P = 0.001) (for hCS n = 8 and for hOLS n = 9 RNA samples from spheroids derived from 4 hiPS cell lines in 1–4 differentiation experiments; see Supplementary Table 1). f–i, Immunostaining of MAP2 at day 54 (f) and GFAP at day 110 (h) and quantification of MAP2+ (g) (two-tailed t-test, t = 3.82, d.f. = 7, **P = 0.006) and GFAP+ (i) (two-tailed t-test, t = 4.615, d.f. = 7, **P = 0.002) at day 54 and day 110 in dissociated hOLS (n = 5 samples each consisting of 4–6 hOLS derived from 3 hiPS cell lines; hiPS cell lines shown in different colors; see Supplementary Table 1). j, Relative expression of VGLUT1 (SLC17A7) (two-tailed Mann–Whitney test, ***P <0.0003) and GAD1 (two-tailed t-test, t = 2.92, d.f. = 17, **P = 0.009), in hCS and hOLS (normalized to GAPDH), as determined by qPCR at day 100 of in vitro culture (for hCS n = 8 and for hOLS n = 11 RNA samples from spheroids derived from 7 hiPS cell lines in 1–3 differentiation experiments; see Supplementary Table 1). k, Day 100 immunostaining of hOLS cryosections for GABA and MAP2. Immunostainings were repeated on hOLS from six independent inductions with similar results. Data are mean ± s.e.m. Scale bars, 50 μm (c, f, h, k upper panel), and 10 μm (k lower panel).
Supplementary Figure 2 Single cell characterization of hOLS.
a, Source of cells in the cluster in Fig. 2b. b, Single cell gene expression pattern of the markers for astrocytes (SOX9), endothelial cells (FLT1), neurons (STMN2), and myeloid cells (CX3CR1). c, Unsupervised hierarchical clustering of all single cells, colored by source. d, Relative gene expression (normalized by GAPDH) as determined by qPCR of MKI67, TOP2A, PDGFRA, and MBP in pooled cDNA from 50 single cells from the proliferating cells, OPCs, NFOs, and myelinating oligodendrocytes clusters (Fig. 2e-g). e, Percentage of cells in the proliferating cells, OPCs, NFOs, and myelinating oligodendrocytes subclusters in two hiPS cell lines. f, Pearson correlation values between the log normalized gene expression data for each oligodendrocyte subcluster between two hiPS cell lines.
Supplementary Figure 3 Primary and hOLS-derived oligodendrocyte differential gene expression and pattern of expression of disease-related genes.
a, Genes that were enriched in oligodendrocyte lineage cells isolated from primary tissue versus hOLS. b, Expression of the housekeeping gene GAPDH across pseudotime (colored by tissue of origin; log2 data normalized by gene). c, Single cell gene expression pattern of disease-implicated genes ARSA, RNASEH2A, and GALC in hOLS, hCS, and primary samples.
Supplementary Figure 4 Electrophysiological characterization of hOLS and myelination.
a, Quantification of the capacitance in bipolar and multipolar Sox10-MCS5::eGFP+ cells (n = 12 bipolar cells, n = 13 multipolar cells, two-tailed Mann–Whitney test, ***P = 0.0006). b, Quantification of the input resistance in bipolar and multipolar Sox10-MCS5::eGFP+ cells (n = 12 bipolar cells, n = 13 multipolar cells, two-tailed Mann–Whitney test, *P = 0.01). For a and b, dots represent individual cells, box edges represent s.e.m., the middle horizontal lines within the box represent the mean, and whiskers represent the 10th and 90th percentiles of the population. c, (upper left) Voltage clamp recording of a Sox10-MCS5::eGFP+ cell showing lack of inward current generation following electrical stimulation indicated by the red dot. (upper right) Voltage clamp recording of Sox10-MCS5::eGFP+ cell showing holding current variance in response to TTX (1 μM) and after treatment with NBQX and APC (lower right). Recordings were repeated in five cells from two independent inductions with similar results. d, Example images of interactions between MBP+ cells and NF-H+ processes in cryosections at days 150–158 of in vitro culture imaged by confocal microscopy. The first and seconds panels of each row are maximum projections, the third panel of each row is an individual z-section, and the right most panels are cross-sections. Immunostainings were repeated on hOLS from three independent inductions with similar results. e,f, Transmission electron microscopy images of hOLS from 8858-3 (e) and 0524-1 (f) at days 150–170 of differentiation showing stages of myelination. Electron microscopy was repeated on hOLS from three independent inductions with similar results. Scale bars, 1 μm (e, f), 50 μm (d left panel), and 10 μm (d middle panel).
Supplementary information
Supplementary Text and Figures
Supplementary Figs. 1–4 and Supplementary Tables 1–3.
Rights and permissions
About this article
Cite this article
Marton, R.M., Miura, Y., Sloan, S.A. et al. Differentiation and maturation of oligodendrocytes in human three-dimensional neural cultures. Nat Neurosci 22, 484–491 (2019). https://doi.org/10.1038/s41593-018-0316-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41593-018-0316-9
This article is cited by
-
Gliomas: a reflection of temporal gliogenic principles
Communications Biology (2024)
-
Human neuronal maturation comes of age: cellular mechanisms and species differences
Nature Reviews Neuroscience (2024)
-
Human cortical spheroids with a high diversity of innately developing brain cell types
Stem Cell Research & Therapy (2023)
-
A beginner’s guide on the use of brain organoids for neuroscientists: a systematic review
Stem Cell Research & Therapy (2023)
-
Potential use of iPSCs for disease modeling, drug screening, and cell-based therapy for Alzheimer’s disease
Cellular & Molecular Biology Letters (2023)