Abstract
Transposable elements, known colloquially as ‘jumping genes’, constitute approximately 45% of the human genome. Cells utilize epigenetic defenses to limit transposable element jumping, including formation of silencing heterochromatin and generation of piwi-interacting RNAs (piRNAs), small RNAs that facilitate clearance of transposable element transcripts. Here we utilize Drosophila melanogaster and postmortem human brain samples to identify transposable element dysregulation as a key mediator of neuronal death in tauopathies, a group of neurodegenerative disorders that are pathologically characterized by deposits of tau protein in the brain. Mechanistically, we find that heterochromatin decondensation and reduction of piwi and piRNAs drive transposable element dysregulation in tauopathy. We further report a significant increase in transcripts of the endogenous retrovirus class of transposable elements in human Alzheimer’s disease and progressive supranuclear palsy, suggesting that transposable element dysregulation is conserved in human tauopathy. Taken together, our data identify heterochromatin decondensation, piwi and piRNA depletion and consequent transposable element dysregulation as a pharmacologically targetable, mechanistic driver of neurodegeneration in tauopathy.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Chuong, E. B., Elde, N. C. & Feschotte, C. Regulatory activities of transposable elements: from conflicts to benefits. Nat. Rev. Genet. 18, 71–86 (2017).
Slotkin, R. K. & Martienssen, R. Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 8, 272–285 (2007).
Brennecke, J. et al. Discrete small RNA-generating loci as master regulators of transposon activity in D rosophila. Cell 128, 1089–1103 (2007).
Li, W. et al. Activation of transposable elements during aging and neuronal decline in D rosophila. Nat. Neurosci. 16, 529–531 (2013).
Wood, J. G. et al. Chromatin-modifying genetic interventions suppress age-associated transposable element activation and extend life span in Drosophila. Proc. Natl. Acad. Sci. USA 113, 11277–11282 (2016).
Burns, K. H. Transposable elements in cancer. Nat. Rev. Cancer 17, 415–424 (2017).
Li, W., Jin, Y., Prazak, L., Hammell, M. & Dubnau, J. Transposable elements in TDP-43-mediated neurodegenerative disorders. PLoS One 7, e44099 (2012).
Li, W. et al. Human endogenous retrovirus-K contributes to motor neuron disease. Sci. Transl. Med. 7, 307ra153 (2015).
Krug, L. et al. Retrotransposon activation contributes to neurodegeneration in a Drosophila TDP-43 model of ALS. PLoS Genet. 13, e1006635 (2017).
Frost, B., Hemberg, M., Lewis, J. & Feany, M. B. Tau promotes neurodegeneration through global chromatin relaxation. Nat. Neurosci. 17, 357–366 (2014).
Wittmann, C. W. et al. Tauopathy in Drosophila: neurodegeneration without neurofibrillary tangles. Science 293, 711–714 (2001).
Hutton, M. et al. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature 393, 702–705 (1998).
Steinhilb, M. L., Dias-Santagata, D., Fulga, T. A., Felch, D. L. & Feany, M. B. Tau phosphorylation sites work in concert to promote neurotoxicity in vivo. Mol. Biol. Cell 18, 5060–5068 (2007).
Dias-Santagata, D., Fulga, T. A., Duttaroy, A. & Feany, M. B. Oxidative stress mediates tau-induced neurodegeneration in Drosophila. J. Clin. Invest. 117, 236–245 (2007).
Khurana, V. et al. A neuroprotective role for the DNA damage checkpoint in tauopathy. Aging Cell 11, 360–362 (2012).
Frost, B., Bardai, F. H. & Feany, M. B. Lamin dysfunction mediates neurodegeneration in tauopathies. Curr. Biol. 26, 129–136 (2016).
Merlo, P. et al. p53 prevents neurodegeneration by regulating synaptic genes. Proc. Natl. Acad. Sci. USA 111, 18055–18060 (2014).
Khurana, V. et al. TOR-mediated cell-cycle activation causes neurodegeneration in a Drosophila tauopathy model. Curr. Biol. 16, 230–241 (2006).
Geiss, G. K. et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat. Biotechnol. 26, 317–325 (2008).
Bardai, F. H. et al. A conserved cytoskeletal signaling cascade mediates neurotoxicity of FTDP-17 tau mutations in vivo. J. Neurosci. 38, 108–119 (2018).
Reilly, M. T., Faulkner, G. J., Dubnau, J., Ponomarev, I. & Gage, F. H. The role of transposable elements in health and diseases of the central nervous system. J. Neurosci. 33, 17577–17586 (2013).
Muotri, A. R. et al. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature 435, 903–910 (2005).
Pélisson, A. et al. About the origin of retroviruses and the co-evolution of the gypsy retrovirus with the Drosophila flamenco host gene. Genetica 100, 29–37 (1997).
Mével-Ninio, M., Pelisson, A., Kinder, J., Campos, A. R. & Bucheton, A. The flamenco locus controls the gypsy and ZAM retroviruses and is required for Drosophila oogenesis. Genetics 175, 1615–1624 (2007).
Frost, B., Götz, J. & Feany, M. B. Connecting the dots between tau dysfunction and neurodegeneration. Trends Cell Biol. 25, 46–53 (2015).
Sarot, E., Payen-Groschêne, G., Bucheton, A. & Pélisson, A. Evidence for a piwi-dependent RNA silencing of the gypsy endogenous retrovirus by the Drosophila melanogaster flamenco gene. Genetics 166, 1313–1321 (2004).
Roy, J., Sarkar, A., Parida, S., Ghosh, Z. & Mallick, B. Small RNA sequencing revealed dysregulated piRNAs in Alzheimer’s disease and their probable role in pathogenesis. Mol. Biosyst. 13, 565–576 (2017).
Qiu, W. et al. Transcriptome-wide piRNA profiling in human brains of Alzheimer’s disease. Neurobiol. Aging 57, 170–177 (2017).
Siomi, M. C., Sato, K., Pezic, D. & Aravin, A. A. PIWI-interacting small RNAs: the vanguard of genome defence. Nat. Rev. Mol. Cell Biol. 12, 246–258 (2011).
Ghildiyal, M. et al. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science 320, 1077–1081 (2008).
Lee, E. J. et al. Identification of piRNAs in the central nervous system. RNA 17, 1090–1099 (2011).
Zamparini, A. L. et al. Vreteno, a gonad-specific protein, is essential for germline development and primary piRNA biogenesis in D rosophila. Development 138, 4039–4050 (2011).
Eissenberg, J. C., Morris, G. D., Reuter, G. & Hartnett, T. The heterochromatin-associated protein HP-1 is an essential protein in Drosophila with dosage-dependent effects on position-effect variegation. Genetics 131, 345–352 (1992).
Reuter, G., Dorn, R., Wustmann, G., Friede, B. & Rauh, G. Third chromosome suppressor of position-effect variegation loci in Drosophila melanogaster. Mol. Gen. Genet. 202, 481–487 (1986).
Fedoroff, N. V. Presidential address. Transposable elements, epigenetics, and genome evolution. Science 338, 758–767 (2012).
Longo, V. D. et al. Interventions to slow aging in humans: are we ready? Aging Cell 14, 497–510 (2015).
Sultana, T., Zamborlini, A., Cristofari, G. & Lesage, P. Integration site selection by retroviruses and transposable elements in eukaryotes. Nat. Rev. Genet. 18, 292–308 (2017).
Coates, J. A. et al. (–)-2′-Deoxy-3′-thiacytidine is a potent, highly selective inhibitor of human immunodeficiency virus type 1 and type 2 replication in vitro. Antimicrob. Agents Chemother. 36, 733–739 (1992).
Andersen, P. R., Tirian, L., Vunjak, M. & Brennecke, J. A heterochromatin-dependent transcription machinery drives piRNA expression. Nature 549, 54–59 (2017).
Brower-Toland, B. et al. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev. 21, 2300–2311 (2007).
Klattenhoff, C. et al. The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell 138, 1137–1149 (2009).
Gonzalez-Cao, M. et al. Human endogenous retroviruses and cancer. Cancer Biol. Med. 13, 483–488 (2016).
Tyagi, R., Li, W., Parades, D., Bianchet, M. A. & Nath, A. Inhibition of human endogenous retrovirus-K by antiretroviral drugs. Retrovirology 14, 21 (2017).
Chuong, E. B., Elde, N. C. & Feschotte, C. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science 351, 1083–1087 (2016).
Heneka, M. T., Golenbock, D. T. & Latz, E. Innate immunity in Alzheimer’s disease. Nat. Immunol. 16, 229–236 (2015).
Fischer, J. A., Giniger, E., Maniatis, T. & Ptashne, M. GAL4 activates transcription in Drosophila. Nature 332, 853–856 (1988).
Dietzl, G. et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448, 151–156 (2007).
Wickersheim, M. L. & Blumenstiel, J. P. Terminator oligo blocking efficiently eliminates rRNA from Drosophila small RNA sequencing libraries. Biotechniques 55, 269–272 (2013).
Kopylova, E., Noé, L. & Touzet, H. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 28, 3211–3217 (2012).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Gramates, L. S. et al. FlyBase at 25: looking to the future. Nucleic Acids Res. 45(D1), D663–D671 (2017).
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A. & Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Kinsella, R. J. et al. Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database (Oxford) 2011, bar030 (2011).
Foroushani, A. et al. Large-scale gene network analysis reveals the significance of extracellular matrix pathway and homeobox genes in acute myeloid leukemia: an introduction to the Pigengene package and its applications. BMC Med. Genomics 10, 16 (2017).
R Development Core Team. R: A language and environment for statistical computing. http://www.R-project.org/(2010).
Wagner, G. P., Kin, K. & Lynch, V. J. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 131, 281–285 (2012).
Sai Lakshmi, S. & Agrawal, S. piRNABank: a web resource on classified and clustered Piwi-interacting RNAs. Nucleic Acids Res. 36, D173–D177 (2008).
Allen, M. et al. Human whole genome genotype and transcriptome data for Alzheimer’s and other neurodegenerative diseases. Sci. Data 3, 160089 (2016).
Hodes, R. J. & Buckholtz, N. Accelerating Medicines Partnership: Alzheimer’s Disease (AMP-AD) Knowledge Portal aids Alzheimer’s drug discovery through open data sharing. Expert Opin. Ther. Targets 20, 389–391 (2016).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Price, A. L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006).
Schroeder, A. et al. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol. Biol. 7, 3 (2006).
Bao, W., Kojima, K. K. & Kohany, O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 6, 11 (2015).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis 2nd edn, Springer International, Basel, Switzerland (2016).
Wald, A. Tests of statistical hypotheses concerning several parameters when the number of observations is large. Trans. Am. Math. Soc. 54, 426–482 (1943).
Acknowledgements
We thank J. Dubnau (Stony Brook University) for gypsy-TRAP, R. Lehmann (New York University School of Medicine) for piwiOE, and M. Feany for tauR406W and tauWT Drosophila stocks. We acknowledge the Texas Advanced Computing Center (TACC; http://www.tacc.utexas.edu) at the University of Texas at Austin for providing high-performance computing resources. This study was supported by the National Institute for Neurological Disorders and Stroke (B.F.) and the Owens Foundation (B.F.). The Mayo human RNA-seq study data was led by N. Ertekin-Taner (Mayo Clinic) as part of the multi-PI U01 AG046139 (MPIs Golde, Ertekin-Taner, Younkin, Price) using samples from the Mayo Clinic Brain Bank. Data collection was supported through funding by NIA grants P50 AG016574, R01 AG032990, U01 AG046139, R01 AG018023, U01 AG006576, U01 AG006786, R01 AG025711, R01 AG017216 and R01 AG003949, NINDS grant R01 NS080820 and the CurePSP Foundation, and support from the Mayo Foundation.
Author information
Authors and Affiliations
Contributions
W.S. and B.F. conceived the study and analyzed the data. W.S. and M.G. performed experiments. H.S. and H.Z. performed RNA-seq analysis. W.S., H.Z. and B.F. contributed to writing the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
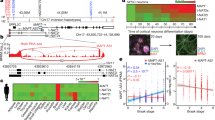
Supplementary Figure 1 Unscaled heatmaps: differentially expressed transposable element transcripts in control versus tauR406W transgenic Drosophila based on RNA-seq.
a, Data presented in Fig. 1a, presented here as raw Transcripts Per Million (TPM). Transposable element transcripts that are differentially expressed in tauR406W transgenic Drosophila heads versus control based on RNA-seq (two-sided Wald test, FDR, p<0.01, n = 3 biologically independent replicates, each consisting of RNA pooled from six heads). 50 transposable elements are significantly increased at the transcript level in tau transgenic Drosophila compared to controls, and 60 transposable elements are significantly decreased. Transcript levels of the majority of these transposable elements are above 20 TPM in at least one of the two conditions. Supplementary Table 2 includes TPM values. b, Fold change measurements in RNA-seq vs. NanoString analyses. Each differentially-expressed transposable element is represented by a dot, where the colors indicate the direction of deregulation as determined based on RNA-Seq data (x-axis, n = 3 biologically independent replicates). The y-axis shows the average of logarithm of fold changes measured using NanoString. The Pearson correlation between the logarithm of fold changes is 0.71 (P<10-7), n = 6 biologically independent replicates. All flies are 10 days old. Full genotypes are listed in Supplementary Table 1
Supplementary Figure 2 Transposable element transcript levels in tauWT transgenic Drosophila.
a, Equivalent levels of transgenic tau protein in heads of control, tauWT, and tauR406W transgenic Drosophila based on western blotting (unpaired, two-tailed Student’s t-test, n.s. = not significant, n = 6 animals per genotype). Western blot is cropped, the full blot is presented in Supplementary Fig. 10. b, c, Transposable element transcript levels in heads of tauWT transgenic Drosophila assayed by NanoString. Transcripts that were identified as increased by RNA-seq in tauR406W transgenic Drosophila are shown in (b), and transcripts that were identified as decreased by RNA-seq in tauR406W transgenic Drosophila are shown in (c) (unpaired, two-tailed Student’s t-test, *P<0.05; **P<0.01; ***P<0.001, n = 6 biologically independent replicates each consisting of RNA pooled from 6 heads, values are relative to control, which was set to 1). Values are mean ± s.e.m. All flies are 10 days old. Full genotypes are listed in Supplementary Table 1. Transposable elements recognized by “generic” probes are listed in Supplementary Table 4
Supplementary Figure 3 Transposable element transcript levels in heads of Drosophila harboring a loss of function mutation in the flamenco locus.
a, Western blot showing equivalent levels of transgenic tau protein in heads of tauR406W transgenic Drosophila harboring loss of function mutations in flamenco (flam) (unpaired, two-tailed Student’s t-test, n.s. = not significant, n = 6 animals per genotype). Western blot is cropped, the full blot is presented in Supplementary Fig. 10. b, c, Transposable element transcript levels in heads of Drosophila harboring a heterozygous loss of function mutation in the flamenco locus assayed by NanoString. Transcripts identified as increased by RNA-seq in tauR406W transgenic Drosophila are shown in (b), and transcripts that were identified as decreased by RNA-seq in tauR406W transgenic Drosophila are shown in (c). d, e, Transposable element transcript levels in heads of Drosophila harboring a homozygous loss of function mutation in the flamenco locus assayed by NanoString. Transcripts identified as increased by RNA-seq in tauR406W transgenic Drosophila are shown in (d), and transcripts that were identified as decreased by RNA-seq in tauR406W transgenic Drosophila are shown in (e). Unpaired, two-tailed Student’s t-test, *P<0.05; **P<0.01; ***P<0.001. For b-e, n = 6 biologically independent replicates each consisting of RNA pooled from 6 heads, values are relative to control, which was set to 1. Values are mean ± s.e.m. All flies are 10 days old. Full genotypes are listed in Supplementary Table 1. Transposable elements recognized by “generic” probes are listed in Supplementary Table 4
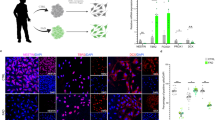
Supplementary Figure 4 Genetic manipulation of piwi.
a, Total levels of piwi protein in whole head homogenates from Drosophila harboring RNAi transgenes targeting piwi (one-way ANOVA with Tukey’s multiple comparison test, **P<0.01, n = 6 animals per genotype). b, Cell cycle activation assayed by PCNA staining in brains of Drosophila harboring RNAi transgenes targeting piwi (one-way ANOVA with Tukey’s multiple comparison test, ****P<0.0001, n = 20 animals per genotype). c, Total transgenic tau levels in whole head homogenates from tauR406W transgenic Drosophila with and without pan-neuronal overexpression of piwi (unpaired, two-tailed Student’s t-test, n.s. = not significant, n = 6 animals per genotype). Values are mean ± s.e.m. All flies are 10 days old. Western blots are cropped in a and c, full blots are presented in Supplementary Fig. 10. Full genotypes are listed in Supplementary Table 1
Supplementary Figure 5 Heterochromatin decondensation increases levels of transposable element transcripts in Drosophila heads.
NanoString analysis of transposable element transcript levels in Drosophila harboring a loss of function mutation in Su(var)205 shown in a, or Su(var)3-9 shown in b (unpaired, two-tailed Student’s t-test, *P<0.05, **P<0.01, ***P<0.001, n = 6 biologically independent replicates each consisting of RNA pooled from 6 heads, values are relative to control, which was set to 1). Values are mean ± s.e.m. All flies are 10 days old. Full genotypes are listed in Supplementary Table 1. Transposable elements recognized by “generic” probes are listed in Supplementary Table 4
Supplementary Figure 6 Transposable element transcript levels in tau transgenic Drosophila in response to dietary restriction.
Transposable element transcript levels in tauR406W transgenic Drosophila with 66% dietary restriction assayed by NanoString (unpaired, two-tailed Student’s t-test, *P<0.05, **P<0.01, n = 6 biologically independent replicates each consisting of RNA pooled from 6 heads, values are relative to tauR406W fed a standard diet, which was set to 1). Values are mean ± s.e.m. All flies are 10 days old. Full genotypes are listed in Supplementary Table 1. Transposable elements recognized by “generic” probes are listed in Supplementary Table 4
Supplementary Figure 7 Dose-response of 3TC treatment.
a, Neuronal death assayed by TUNEL in tauR406W transgenic Drosophila treated with the indicated dose of 3TC (unpaired, two-tailed Student’s t-test, *P = 0.0157, ****P<0.0001, n = 6 animals per treatment). b, Locomotor activity in control and tauR406W transgenic Drosophila treated with the indicated dose of 3TC (one-way ANOVA with Tukey’s multiple comparison test, n.s. = not significant, *P<0.05, **P = 0.0077, ****P<0.0001, n = 18 animals per genotype, per treatment). Values are mean ± s.e.m. All flies are 10 days old. Full genotypes listed in Supplementary Table 1
Supplementary Figure 8 Transposable element transcript levels in cerebellum of human tauopathy.
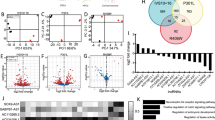
Heatmaps reflecting fold change of differentially expressed transposable element transcripts in human cerebellum in a, control versus Alzheimer’s disease (AD), and b, control versus progressive supranuclear palsy (PSP), based on RNA-seq (two-sided Wald test, FDR, P<0.01). c, Differentially expressed transposable elements in postmortem Alzheimer’s disease and progressive supranuclear palsy cerebellum. HERVs are significantly over-represented among transposable elements that are increased in tauopathy (hypergeometric test, adjusted P = 0.0003). Non-LTR are significantly over-represented among transposable elements that are decreased in tauopathy (hypergeometric test, adjusted P = 0.01). d, Principal component analyses of differentially expressed transposable elements in control, Alzheimer’s disease, and progressive supranuclear palsy cerebellum (Kolmogorov-Smirnov test, P<10-14). e, Violin plots show that control samples are relatively farther from the center of the cluster, defined as the median of tauopathy cerebellum samples. Euclidian distance was computed using the two first principal components of those transposable elements that are differentially expressed among the three conditions. For control, AD, and PSP respectively, minima = 0.5, 0.08, 0.3, maxima = 10.4, 12.7, 6.9, median = 3.1, 2.5, 2.4, mean = 4.0, 3.4, 2.6, 1st quantile = 2.7, 1.4, 1.4, 3rd quantile = 4.6, 4.2, 3.5. Control n = 25, Alzheimer’s disease n = 76, progressive supranuclear palsy n = 78 biologically independent replicates in a-e
Supplementary Figure 9 Graphical summary.
Pathological tau disrupts heterochromatin- and piRNA-mediated transposable element silencing. Activation of transposable elements promotes aberrant activation of the cell cycle in post-mitotic neurons, which is sufficient to induce neuronal death. A nucleoside analog inhibitor of reverse transcriptase, 3TC, as well as dietary restriction suppress transposable element dysregulation in tauopathy, and decrease tau-induced neurotoxicity
Supplementary information
Supplementary Text, Figures and Tables
Supplementary Figures 1–10, Supplementary Tables 1, 4 and 8
Supplementary Table 2 - Differential expression analysis of transposable elements in Drosophila based on RNA-seq.
The first column includes the FlyBase IDs of transposable elements, which are ordered on the basis of P-value.The next six columns, computed using DESeq2, are the mean of the normalized counts (basemean); the logarithm of the fold change (log2FoldChange); the standard error of the log2FoldChange (lfcSE); the two-sided Wald statistic,defined as the log2FoldChange divided by lfcSE (stat); the P-value of the Wald test; and the Benjamini–Hochberg adjusted P-value (padj). The raw counts from RNA-seq are also included
Supplementary Table 3
Sequences of NanoString probes
Supplementary Table 5 - Differential expression analysis of piRNAs in Drosophila.
The first column is the piRNA name sorted by P-value (two-sided Wald test), and the next six columns are similar to those in Supplementary Table 2. In addition to raw counts, the sum of counts in each condition is included, as well as the ratio of the sum of the counts in tau transgenic samples over the sum of the counts in control samples. Because a piRNA can have multiple copies in the Drosophila genome, we included the number of copies (frequency) and their genomic locations, which are separated by an asterisk. The last column is the sequence of the piRNA
Supplementary Table 6 - Genomic locations of Drosophila piRNAs.
The first column is the piRNA name sorted by P-value of two-sided tests. The chromosome, start, end, strand, sequence, number of copies (frequency), and adjusted P-value from the small RNA-seq data are included. n = 4 biologically independent replicates of each genotype, each consisting of RNA pooled from 6 heads. P-values were calculated on the basis of a two-sided Wald test, FDR
Supplementary Table 7 - Differential expression of transposable elements in human tauopathy.
In each of the first six sheets, the transposable elements are sorted on the basis of the P-value corresponding to the tissue (cortex or cerebellum) and the condition specified in the sheet name. Columns are similar to those in Supplementary Table 2. The last two sheets are the clinical data, including the diagnosis and the RIN of RNA samples. More detailed clinical information, including gender, age at death, apolipoprotein E (ApoE), sequencing flow cell batch, and post mortem interval, is available at the Synapse.org website (cortex, http://dx.doi.org/10.7303/syn5223705.3; cerebellum, http://dx.doi.org/10.7303/syn3817650.6). P-values were calculated on the basis of a two-sided Wald test, FDR
Supplementary Table 9
Statistical analyses
Rights and permissions
About this article
Cite this article
Sun, W., Samimi, H., Gamez, M. et al. Pathogenic tau-induced piRNA depletion promotes neuronal death through transposable element dysregulation in neurodegenerative tauopathies. Nat Neurosci 21, 1038–1048 (2018). https://doi.org/10.1038/s41593-018-0194-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41593-018-0194-1
This article is cited by
-
The interaction between ageing and Alzheimer's disease: insights from the hallmarks of ageing
Translational Neurodegeneration (2024)
-
Condensin-mediated restriction of retrotransposable elements facilitates brain development in Drosophila melanogaster
Nature Communications (2024)
-
Activation of human endogenous retroviruses and its physiological consequences
Nature Reviews Molecular Cell Biology (2024)
-
Phosphorylation regulates tau’s phase separation behavior and interactions with chromatin
Communications Biology (2024)
-
Alterations of mRNAs and Non-coding RNAs Associated with Neuroinflammation in Alzheimer’s Disease
Molecular Neurobiology (2024)