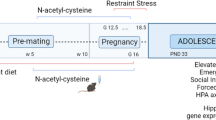
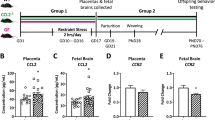
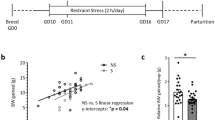
Abstract
Early prenatal stress disrupts maternal-to-offspring microbiota transmission and has lasting effects on metabolism, physiology, cognition, and behavior in male mice. Here we show that transplantation of maternal vaginal microbiota from stressed dams into naive pups delivered by cesarean section had effects that partly resembled those seen in prenatally stressed males. However, transplantation of control maternal vaginal microbiota into prenatally stressed pups delivered by cesarean section did not rescue the prenatal-stress phenotype. Prenatal stress was associated with alterations in the fetal intestinal transcriptome and niche, as well as with changes in the adult gut that were altered by additional stress exposure in adulthood. Further, maternal vaginal transfer also partially mediated the effects of prenatal stress on hypothalamic gene expression, as observed after chronic stress in adulthood. These findings suggest that the maternal vaginal microbiota contribute to the lasting effects of prenatal stress on gut and hypothalamus in male mice.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Bale, T. L. et al. Early life programming and neurodevelopmental disorders. Biol. Psychiatry 68, 314–319 (2010).
Cryan, J. F. & Dinan, T. G. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat. Rev. Neurosci. 13, 701–712 (2012).
Sampson, T. R. & Mazmanian, S. K. Control of brain development, function, and behavior by the microbiome. Cell Host Microbe 17, 565–576 (2015).
Gur, T. L. et al. Prenatal stress affects placental cytokines and neurotrophins, commensal microbes, and anxiety-like behavior in adult female offspring. Brain Behav. Immun. 64, 50–58 (2017).
Golubeva, A. V. et al. Prenatal stress-induced alterations in major physiological systems correlate with gut microbiota composition in adulthood. Psychoneuroendocrinology 60, 58–74 (2015).
Zijlmans, M. A., Korpela, K., Riksen-Walraven, J. M., de Vos, W. M. & de Weerth, C. Maternal prenatal stress is associated with the infant intestinal microbiota. Psychoneuroendocrinology 53, 233–245 (2015).
Jašarević, E., Morrison, K. E. & Bale, T. L. Sex differences in the gut microbiome-brain axis across the lifespan. Phil. Trans. R. Soc. Lond. B 371, 20150122 (2016).
Yatsunenko, T. et al. Human gut microbiome viewed across age and geography. Nature 486, 222–227 (2012).
Borre, Y. E. et al. Microbiota and neurodevelopmental windows: implications for brain disorders. Trends Mol. Med. 20, 509–518 (2014).
Jašarević, E., Howard, C. D., Misic, A. M., Beiting, D. P. & Bale, T. L. Stress during pregnancy alters temporal and spatial dynamics of the maternal and offspring microbiome in a sex-specific manner. Sci. Rep. 7, 44182 (2017).
Jašarević, E., Howerton, C. L., Howard, C. D. & Bale, T. L. Alterations in the vaginal microbiome by maternal stress are associated with metabolic reprogramming of the offspring gut and brain. Endocrinology 156, 3265–3276 (2015).
Bronson, S. L. & Bale, T. L. Prenatal stress-induced increases in placental inflammation and offspring hyperactivity are male-specific and ameliorated by maternal antiinflammatory treatment. Endocrinology 155, 2635–2646 (2014).
Mueller, B. R. & Bale, T. L. Impact of prenatal stress on long term body weight is dependent on timing and maternal sensitivity. Physiol. Behav. 88, 605–614 (2006).
Mueller, B. R. & Bale, T. L. Early prenatal stress impact on coping strategies and learning performance is sex dependent. Physiol. Behav. 91, 55–65 (2007).
Mueller, B. R. & Bale, T. L. Sex-specific programming of offspring emotionality after stress early in pregnancy. J. Neurosci. 28, 9055–9065 (2008).
Pankevich, D. E., Mueller, B. R., Brockel, B. & Bale, T. L. Prenatal stress programming of offspring feeding behavior and energy balance begins early in pregnancy. Physiol. Behav. 98, 94–102 (2009).
Dominguez-Bello, M. G. et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl Acad. Sci. USA 107, 11971–11975 (2010).
Dominguez-Bello, M. G. et al. Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer. Nat. Med. 22, 250–253 (2016).
Ericsson, A. C. & Franklin, C. L. Manipulating the gut microbiota: methods and challenges. ILAR J. 56, 205–217 (2015).
Bravo, J. A. et al. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc. Natl Acad. Sci. USA 108, 16050–16055 (2011).
Buffington, S. A. et al. Microbial reconstitution reverses maternal diet-induced social and synaptic deficits in offspring. Cell 165, 1762–1775 (2016).
Ait-Belgnaoui, A. et al. Prevention of gut leakiness by a probiotic treatment leads to attenuated HPA response to an acute psychological stress in rats. Psychoneuroendocrinology 37, 1885–1895 (2012).
Ait-Belgnaoui, A. et al. Probiotic gut effect prevents the chronic psychological stress-induced brain activity abnormality in mice. Neurogastroenterol. Motil. 26, 510–520 (2014).
Wells, J. M. Immunomodulatory mechanisms of lactobacilli. Microb. Cell Fact. 10(Suppl 1), S17 (2011).
Makarova, K. et al. Comparative genomics of the lactic acid bacteria. Proc. Natl Acad. Sci. USA 103, 15611–15616 (2006).
Almirón, M., Traglia, G., Rubio, A. & Sanjuan, N. Colonization of the mouse upper gastrointestinal tract by lactobacillus murinus: a histological, immunocytochemical, and ultrastructural study. Curr. Microbiol. 67, 395–398 (2013).
Howerton, C. L., & Bale, T. L. Targeted placental deletion of OGT recapitulates the prenatal stress phenotype including hypothalamic mitochondrial dysfunction. Proc. Natl Acad. Sci. USA 111, 9639–9644 (2014).
Rawls, J. F., Mahowald, M. A., Ley, R. E. & Gordon, J. I. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell 127, 423–433 (2006).
Akira, S. & Takeda, K. Toll-like receptor signalling. Nat. Rev. Immunol. 4, 499–511 (2004).
Deshmukh, H. S. et al. The microbiota regulates neutrophil homeostasis and host resistance to Escherichia coli K1 sepsis in neonatal mice. Nat. Med. 20, 524–530 (2014).
Molloy, M. J. et al. Intraluminal containment of commensal outgrowth in the gut during infection-induced dysbiosis. Cell Host Microbe 14, 318–328 (2013).
Rakoff-Nahoum, S. et al. Analysis of gene-environment interactions in postnatal development of the mammalian intestine. Proc. Natl Acad. Sci. USA 112, 1929–1936 (2015).
Cani, P. D. et al. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes 57, 1470–1481 (2008).
Seo, S. U. et al. Distinct commensals induce interleukin-1β via NLRP3 inflammasome in inflammatory monocytes to promote intestinal inflammation in response to injury. Immunity 42, 744–755 (2015).
Nemeghaire, S. et al. The ecological importance of the Staphylococcus sciuri species group as a reservoir for resistance and virulence genes. Vet. Microbiol. 171, 342–356 (2014).
Ahrne, S. & Hagslatt, M. L. Effect of lactobacilli on paracellular permeability in the gut. Nutrients 3, 104–117 (2011).
Houghteling, P. D. & Walker, W. A. Why is initial bacterial colonization of the intestine important to infants’ and children’s health? J. Pediatr. Gastroenterol. Nutr. 60, 294–307 (2015).
Bailey, M. T. & Coe, C. L. Maternal separation disrupts the integrity of the intestinal microflora in infant rhesus monkeys. Dev. Psychobiol. 35, 146–155 (1999).
Bailey, M. T., Lubach, G. R. & Coe, C. L. Prenatal stress alters bacterial colonization of the gut in infant monkeys. J. Pediatr. Gastroenterol. Nutr. 38, 414–421 (2004).
Winter, S. E. & Bäumler, A. J. Why related bacterial species bloom simultaneously in the gut: principles underlying the ‘Like will to like’ concept. Cell. Microbiol. 16, 179–184 (2014).
Gomez de Agüero, M. et al. The maternal microbiota drives early postnatal innate immune development. Science 351, 1296–1302 (2016).
Mayer, E. A. The neurobiology of stress and gastrointestinal disease. Gut 47, 861–869 (2000).
Mayer, E. A. Gut feelings: the emerging biology of gut-brain communication. Nat. Rev. Neurosci. 12, 453–466 (2011).
Kerschensteiner, M., Stadelmann, C., Dechant, G., Wekerle, H. & Hohlfeld, R. Neurotrophic cross-talk between the nervous and immune systems: implications for neurological diseases. Ann. Neurol. 53, 292–304 (2003).
Nagy, A., Gertsenstein, M., Vintersten, K. & Behringer, R. Caesarean section and fostering. CSH Protocols 2006, pdb.prot4381 (2006).
Murphy, D. Caesarean section and fostering. Methods Mol. Biol. 18, 177–178 (1993).
McEuen, J. G., Semsar, K. A., Lim, M. A. & Bale, T. L. Influence of sex and corticotropin-releasing factor pathways as determinants in serotonin sensitivity. Endocrinology 150, 3709–3716 (2009).
Bale, T. L. et al. Mice deficient for corticotropin-releasing hormone receptor-2 display anxiety-like behaviour and are hypersensitive to stress. Nat. Genet. 24, 410–414 (2000).
Rodgers, A. B., Morgan, C. P., Bronson, S. L., Revello, S. & Bale, T. L. Paternal stress exposure alters sperm microRNA content and reprograms offspring HPA stress axis regulation. J. Neurosci. 33, 9003–9012 (2013).
Kozich, J. J., Westcott, S. L., Baxter, N. T., Highlander, S. K. & Schloss, P. D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 79, 5112–5120 (2013).
Schloss, P. D. et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541 (2009).
Caporaso, J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010).
Paulson, J. N., Stine, O. C., Bravo, H. C. & Pop, M. Differential abundance analysis for microbial marker-gene surveys. Nat. Methods 10, 1200–1202 (2013).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Bray, N. L., Pimentel, H., Melsted, P. & Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 (2016).
Soneson, C., Love, M. I. & Robinson, M. D. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res. 4, 1521 (2015).
Law, C. W., Chen, Y., Shi, W. & Smyth, G. K. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15, R29 (2014).
Schmittgen, T. D. & Livak, K. J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108 (2008).
Plaisier, S. B., Taschereau, R., Wong, J. A. & Graeber, T. G. Rank-rank hypergeometric overlap: identification of statistically significant overlap between gene-expression signatures. Nucleic Acids Res. 38, e169 (2010).
RC Team. R: a Language and Environment for Statistical Computing. (R Foundation for Statistical Computing, Vienna, Austria, 2014).
Acknowledgements
The research reported in this publication was supported by a pilot award from the PennVet Center for Host–Microbial Interactions at the University of Pennsylvania. T.L.B was supported by the National Institutes of Mental Health under Award Numbers P50-MH099910, MH 104184, MH 091258, MH 087597, MH 073030, and MH 108286. E.J. was supported by the National Institutes of Health National Research Service Award F32 MH 109298. T.W. was supported by National Institutes of Health Postdoctoral Research Grant F32 AI 114080.
Author information
Authors and Affiliations
Contributions
E.J. and T.L.B. designed the experiments and wrote the manuscript. E.J. performed the cesarean delivery experiments, molecular biology experiments, and behavioral experiments and performed bioinformatics analysis of RNA-seq data, 16S rRNA marker gene sequencing data, and whole-metagenomic shotgun sequencing data. C.D.H. assisted with all cesarean delivery experiments. A.M.M. and D.P.B. provided bioinformatics support for RNA-seq, 16S rRNA, and shotgun whole-metagenomics. K.E.M. collected PVN brain micropunches for RNA-seq analyses. T.W. conducted fetal intestine cell isolation and flow cytometry analyses. T.W., P.S., and C.A.H. provided resources and equipment for multicolor flow cytometry of fetal intestinal tissues. All authors contributed to manuscript editing and revision.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Restoration of P2 microbiota patterns of cesarean–delivered offspring via the maternal vaginal inoculant.
(a) Taxonomic classifications of microbiota colonizing the PN2 colon of C-section offspring that did not receive a maternal vaginal inoculant (noninoculated), C-section offspring that received a maternal vaginal inoculant (inoculated), and vaginally delivered offspring demonstrating greater similarity in microbiota community composition between inoculated and vaginally delivered offspring. N = 8 Caesarean delivered noninoculated mice; N = 24 Caesarean delivered inoculated mice; N = 29 Vaginally delivered mice. (b) Communities clustered using PCoA of the unweighted UniFrac distance matrix where each point corresponds to a sample collected from PN2 colons of vaginally delivered CTL and prenatal stress-exposed offspring, C-section noninoculated offspring, and C→C, C→S, S→C and S→S offspring. The percentage of variation explained by the PC is indicated on the axes. PCoA, principal coordinates analysis. N = 9 vaginally delivered control males; N = 20 vaginally delivered prenatal-stress exposed males; N = 8 noninoculated C-section delivered males; N = 6 C→C males; N = 9 C→S males; N = 6 S→C males; N = 5 S→S males. (c) Barplot of Unweighted UniFrac Distances showing significant differences in the community structure of noninoculated C-section offspring, vaginally delivered CTL and prenatal stress offspring, C→C, C→S, S→C, and S→S offspring at PN2 (One-way ANOVA, treatment, F6,386 = 9.253, P = 0.00000000175). N = 9 vaginally delivered control males; N = 20 vaginally delivered prenatal-stress exposed males; N = 8 noninoculated C-section delivered males; N = 6 C→C males; N = 9 C→S males; N = 6 S→C males; N = 5 S→S males. Data represented as mean ± SEM with individual UniFrac distance correlations overlaid. (d) Barplot (mean ± SEM) of Unweighted UniFrac Distances showing no significant difference in community structure of C→C, C→S, S→C, and S→S offspring at PN2. N = 6 C→C males; N = 9 C→S males; N = 6 S→C males; N = 5 S→S males. Data represented as mean ± SEM with individual UniFrac distance correlations overlaid. (e) Barplot (mean ± SEM) of the Shannon Diversity Index showing no effect of prenatal stress on community diversity in vaginally delivered offspring. Bacterial diversity is decreased in Noninoculated (NIC) offspring relative to vaginally delivered offspring (Vaginally delivered control vs. NIC, t15 = 2.774, P = 0.0142; Vaginally delivered prenatal stress exposed vs. NIC, t26 = 2.49, P = 0.0195, Unpaired t-Test). No difference in community diversity between vaginally delivered offspring and C-section offspring that received a maternal vaginal inoculant (C→C, C→S, S→C, and S→S offspring) (One-way ANOVA, main effect of treatment, F5,47 = 0.3447, P = 0.8831). N = 9 vaginally delivered control males; N = 20 vaginally delivered prenatal-stress exposed males; N = 8 noninoculated C-section delivered males; N = 6 C→C males; N = 9 C→S males; N = 6 S→C males; N = 5 S→S males. * P < 0.05. Data represented as mean ± SEM with individual data points overlaid.
Supplementary Figure 2 Characterization of the Lactobacillus species in the P2 colon.
(a) Barplot (mean ± SEM) of Lactobacillus species abundance detected in the postnatal day 2 colon showing that L.murinus is the lactobacilli species present in more than half of samples in the postnatal day 2 colon of C→C mice. N = 6 C→C males. Data represented as mean ± SEM with individual data points overlaid. (b-d) No treatment differences in abundance of L. johnsonii, L. animalis, and L. apodemi in the postnatal day 2 gut of C→C, C→S, S→C, and S→S offspring (One-way ANOVA; L. johnsonii, F3,25 = 0.9404, P = 0.4360; L. animalis, F3,25 = 1.210, P = 0.3265; L. adopemi, F3,25 = 1.322, P = 0.2895). N = 6 C→C males; N = 10 C→S males; N = 7 S→C males; N = 6 S→S males. Data represented as mean ± SEM with individual data points overlaid.
Supplementary Figure 3 The maternal vaginal microbiome does not impact hypothalamic–pituitary–adrenal stress axis responsivity in female offspring.
(a) Production of corticosterone following a 15 min restraint stress in vaginally delivered and cross-fostered control and prenatal stress-exposed female offspring is different across time but not by treatment or its interaction (two-way ANOVA, main effect of time, F3,33 = 63.35, P = 0.000000000000088). N = 6 vaginally delivered control females; N = 7 vaginally delivered prenatal stress-exposed females. Data is representative of two experimental replicates. Two-way ANOVA with time as a repeated measure. Data are presented as mean ± range with bars representing min and max. (b) Production of corticosterone following a 15 min restraint stress in C→C, C→S, S→C, and S→S females is different across time but not by treatment or its interaction (two-way ANOVA, main effect of time, F3,24 = 130.7, P = 0.000000000000005). N = 3 C→C females; N = 3 C→S females; N = 3 S→C females; N = 3 S→S females. Data is representative of two experimental replicates. Two-way ANOVA with time as a repeated measure. Data are presented as mean ± range with bars representing min and max. (c) Total Area Under the Curve (AUC) production of corticosterone in females following a 15 min restrain stress. (Left) Total AUC corticosterone levels did not differ between vaginally delivered and cross-fostered control females and prenatal stress-exposed females (Unpaired two-sided t-Test, t11 = 1.013, P = 0.33). (Right) Total AUC corticosterone levels did not differ between C→C, C→S, S→C, and S→S females (one-way ANOVA, F3,8 = 2.346, P = 0.1490). N = 6 vaginally delivered control females; N = 7 vaginally delivered prenatal stress-exposed females; N = 3 C→C females; N = 3 C→S females; N = 3 S→C females; N = 3 S→S females. Data is representative of two experimental replicates. Unpaired two-sided t-Test comparing vaginally delivered and cross-fostered offspring; One-way ANOVA compared cesarean delivered and inoculated offspring. Data are presented as mean ± SEM with individual data points overlaid.
Supplementary Figure 4 Adult locomotion and anxiety-like behavior are not affected by cross-fostering, cesarean delivery, and inoculation of maternal vaginal fluid.
(a-c) No difference in transitions, time in light, and distance travelled in the light-dark box paradigm between vaginally delivered control and prenatal stress-exposed adult males, or between C→C, C→S, S→C, and S→S adult males. Vaginally delivered control and prenatal stress-exposed males were tested separately from C→C, C→S, S→C, and S→S males (Vaginally delivered control vs. prenatal stress exposed males, transitions, t12 = 1.591, P = 0.1376; time in light, t12 = 0.0019, P = 0.999; distance, t12 = 0.37, P = 0.7177, Unpaired two-sided t-Test; Comparison between C→C, C→S, S→C, and S→S males, transitions, F3,12 = 0.106, P = 0.95; time in light, F3,12 = 0.4836, P = 0.701; distance, F3,12 = 0.2604, P = 0.8523, One-way ANOVA). Box-whisker plot center line, median; box limits, upper and lower quartiles; whiskers represent Tukey fences. N = 7 vaginally delivered control males; N = 7 vaginally delivered prenatal-stressed males; N = 4 C→C males; N = 3 C→S males; N = 6 S→C males; N = 3 S→S males. Data represented as box-whisker plot. (d-f) No difference in transitions, time in light, and distance travelled in the light-dark box paradigm between vaginally delivered control and prenatal stress-exposed adult females, or between C→C, C→S, S→C, and S→S adult females. Vaginally delivered control and prenatal stress-exposed females were tested separately from C→C, C→S, S→C, and S→S females. (Vaginally delivered control vs. prenatal stress exposed females, transitions, t12 = 0.5078, P = 0.6208; time in light, t12 = 1.025, P = 0.3255; distance, t12 = 0.0448, P = 0.965, Unpaired two-sided t-Test; Comparison between C→C, C→S, S→C, and S→S females, transitions, F3,8 = 0.7957, P = 0.5299; time in light, F3,8 = 0.7363, P = 0.5592; distance, F3,8 = 0.3216, P = 0.81, One-way ANOVA). Box-whisker plot center line, median; box limits, upper and lower quartiles; whiskers represent Tukey fences. N = 7 vaginally delivered control females; N = 7 vaginally delivered prenatal stress-exposed females; N = 3 C→C females; N = 3 C→S females; N = 3 S→C females; N = 3 S→S females. Data represented as box-whisker plot.
Supplementary Figure 5 Early prenatal stress disrupts sex differences in the fetal intestine at E18.5.
(a) Schematic of experimental design to assess whether early prenatal stress exposure results in sex-specific alterations to the fetal gut. (b) Venn diagram of differential expression analysis of mouse fetal intestinal genes via RNASeq. Circle labels refer to the sex and treatment comparisons: MCTLvFCTL are the comparisons between control males and control females, MCTLvMEPS are the comparisons between control males and prenatal stress-exposed males, and FCTLvFEPS are comparisons between female control and prenatal stress-exposed females. The numbers within the diagram represent the number of genes characterized as differential expression between these groups and the number of differentially expressed genes that overlap between treatment comparisons. FCTL, female control; MCTL, male control; FEPS, female EPS; MEPS, male EPS. EPS, early prenatal stress (linear fit model, statistical parameters: p < 0.01, logFC = 1.5). N = 4 control males; N = 4 control females; N = 5 prenatal-stress exposed females; N = 4 prenatal stress-exposed males. (c) Heatmap depicting mean expression of E18.5 fetal intestinal genes. The heatmaps show significant sex differences between control males and control females that are disrupted in prenatal stress-exposed offspring (linear fit model, statistical parameters: p < 0.01, logFC = 1.5). N = 4 control males; N = 4 control females; N = 5 prenatal-stress exposed females; N = 4 prenatal stress-exposed males. (d) Selected fetal intestinal genes that are differentially expressed between control males and control females but not between prenatally stressed males and prenatally stressed females. The disruption of the sex difference by prenatal stress is due to a more female-typical expression pattern in prenatally stressed males. Y-axis shows fold change relative to expression in control males. Box-whisker plot center line, median; box limits, upper and lower quartiles; whiskers represent Tukey fences. N = 4 control males; N = 4 control females; N = 5 prenatal-stress exposed females; N = 4 prenatal stress-exposed males. Data represented as a box-whisker plots. (e) Prenatal stress increases expression of the pro-inflammatory cytokine tumor necrosis factor-α in the fetal intestine in males but not in females (t7 = 2.624, P = 0.0342). * p < 0.05, Unpaired two-sided t-Test. Box-whisker plot center line, median; box limits, upper and lower quartiles; whiskers represent Tukey fences. N = 4 control males; N = 5 prenatal stress-exposed males; N = 7 control female; N = 7 prenatal stress-exposed females. Data represented as a box-whisker plots.
Supplementary Figure 6 Effects of early prenatal stress on adult immunity following chronic stress in adulthood.
(a) (Left) Representative flow cytometry contour plots showing gating of CD11b + Ly6C + cells and the percentage of Ly6C + (lower circle) and Ly6Chi (upper circle) cells from control and prenatal-stress exposed males following chronic stress exposure. (Right) Analysis of CD45+CD11b+Ly6G-Ly6C+ monocytes in the adult spleen following chronic variable stress exposure revealed main effect of prenatal stress on monocyte populations (Two-way ANOVA, treatment*immune cell marker interaction, F1,12 = 5.310, P = 0.0399; Ly6Chi CTL vs. Prenatal stress, t12 = 2.062, P = 0.0616). N = 3 control males; N = 5 prenatal-stress exposed males. Data represented mean ± SEM with individual data points overlaid. (b) (Left) Representative flow cytometry contour plots showing intracellular staining of TNFa in Ly6C + cells and the percentage from control and prenatal-stress exposed adult males following chronic stress exposure. (Right) Intracellular staining analysis of TNFα in CD45+CD11b+Ly6G-Ly6C+ monocytes from the adult spleen of male mice exposed to chronic variable stress revealed no significant difference in TNFα staining in monocytes between prenatal stress-exposed males and control males (Unpaired t-Test, t4 = 2.659, P = 0.0554). N = 3 control males; N = 5 prenatal-stress exposed males. Data represented mean ± SEM with individual data points overlaid.
Supplementary Information
Supplementary Text and Figures
Supplementary Figures 1–6
Supplementary Table 1
Gene set enrichment analysis comparing embryonic day 18.5 gene expression patterns of animals exposed to early prenatal stress or not revealed significant enrichment in genes involved in the innate immunity in exposed animals.
Supplementary Table 2
Gene set enrichment analysis comparing embryonic day 18.5 gene expression patterns revealing significant enrichment in genes involved in response to interferons.
Supplementary Table 3
Gene set enrichment analysis comparing adult PVN gene expression patterns of C→C, C→S, S→C, S→S males exposed to chronic stress in adulthood, revealing significant enrichment in genes involved butanoate metabolism in stress-exposed C→C males relative to S→S males.
Supplementary Table 4
Gene set enrichment analysis comparing adult PVN gene expression patterns of C→C, C→S, S→C, S→S males exposed to chronic stress in adulthood, revealing significant enrichment in genes involved fatty acid metabolism in stress-exposed C→C males relative to S→S males.
Supplementary Table 5
Gene set enrichment analysis comparing adult PVN gene expression patterns of C→C, C→S, S→C, S→S males exposed to chronic stress in adulthood, revealing significant enrichment in genes involved in neurotrophin signaling in stress-exposed C→C males relative to S→S males.
Rights and permissions
About this article
Cite this article
Jašarević, E., Howard, C.D., Morrison, K. et al. The maternal vaginal microbiome partially mediates the effects of prenatal stress on offspring gut and hypothalamus. Nat Neurosci 21, 1061–1071 (2018). https://doi.org/10.1038/s41593-018-0182-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41593-018-0182-5
This article is cited by
-
Autism spectrum disorders and the gastrointestinal tract: insights into mechanisms and clinical relevance
Nature Reviews Gastroenterology & Hepatology (2024)
-
Extracellular vesicles are dynamic regulators of maternal glucose homeostasis during pregnancy
Scientific Reports (2023)
-
Should we modulate the neonatal microbiome and what should be the goal?
Microbiome (2022)
-
Ontogenetic rules for the molecular diversification of hypothalamic neurons
Nature Reviews Neuroscience (2022)
-
Cinnamaldehyde prevents intergenerational effect of paternal depression in mice via regulating GR/miR-190b/BDNF pathway
Acta Pharmacologica Sinica (2022)