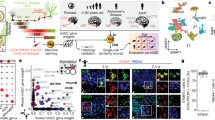
The dentate gyrus of the hippocampus is a brain region in which neurogenesis persists into adulthood; however, the relationship between developmental and adult dentate gyrus neurogenesis has not been examined in detail. Here we used single-cell RNA sequencing to reveal the molecular dynamics and diversity of dentate gyrus cell types in perinatal, juvenile, and adult mice. We found distinct quiescent and proliferating progenitor cell types, linked by transient intermediate states to neuroblast stages and fully mature granule cells. We observed shifts in the molecular identity of quiescent and proliferating radial glia and granule cells during the postnatal period that were then maintained through adult stages. In contrast, intermediate progenitor cells, neuroblasts, and immature granule cells were nearly indistinguishable at all ages. These findings demonstrate the fundamental similarity of postnatal and adult neurogenesis in the hippocampus and pinpoint the early postnatal transformation of radial glia from embryonic progenitors to adult quiescent stem cells.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Amaral, D. G., Scharfman, H. E. & Lavenex, P. The dentate gyrus: fundamental neuroanatomical organization (dentate gyrus for dummies). Prog. Brain Res. 163, 3–22 (2007).
Altman, J. & Bayer, S. A. Migration and distribution of two populations of hippocampal granule cell precursors during the perinatal and postnatal periods. J. Comp. Neurol. 301, 365–381 (1990).
Piatti, V. C., Espósito, M. S. & Schinder, A. F. The timing of neuronal development in adult hippocampal neurogenesis. Neuroscientist 12, 463–468 (2006).
Malatesta, P., Hartfuss, E. & Götz, M. Isolation of radial glial cells by fluorescent-activated cell sorting reveals a neuronal lineage. Development 127, 5253–5263 (2000).
Malatesta, P. & Götz, M. Radial glia - from boring cables to stem cell stars. Development 140, 483–486 (2013).
Kriegstein, A. & Alvarez-Buylla, A. The glial nature of embryonic and adult neural stem cells. Annu. Rev. Neurosci. 32, 149–184 (2009).
Doetsch, F., Caillé, I., Lim, D. A., García-Verdugo, J. M. & Alvarez-Buylla, A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell 97, 703–716 (1999).
Seri, B., García-Verdugo, J. M., McEwen, B. S. & Alvarez-Buylla, A. Astrocytes give rise to new neurons in the adult mammalian hippocampus. J. Neurosci. 21, 7153–7160 (2001).
Steiner, B. et al. Differential regulation of gliogenesis in the context of adult hippocampal neurogenesis in mice. Glia 46, 41–52 (2004).
Garcia, A. D. R., Doan, N. B., Imura, T., Bush, T. G. & Sofroniew, M. V. GFAP-expressing progenitors are the principal source of constitutive neurogenesis in adult mouse forebrain. Nat. Neurosci. 7, 1233–1241 (2004).
Altman, J. & Das, G. D. Autoradiographic and histological evidence of postnatal hippocampal neurogenesis in rats. J. Comp. Neurol. 124, 319–335 (1965).
Kaplan, M. S. & Hinds, J. W. Neurogenesis in the adult rat: electron microscopic analysis of light radioautographs. Science 197, 1092–1094 (1977).
Eriksson, P. S. et al. Neurogenesis in the adult human hippocampus. Nat. Med. 4, 1313–1317 (1998).
Gould, E. et al. Hippocampal neurogenesis in adult Old World primates. Proc. Natl. Acad. Sci. USA 96, 5263–5267 (1999).
Gage, F. H. Mammalian neural stem cells. Science 287, 1433–1438 (2000).
Kempermann, G., Jessberger, S., Steiner, B. & Kronenberg, G. Milestones of neuronal development in the adult hippocampus. Trends Neurosci. 27, 447–452 (2004).
Kempermann, G., Kuhn, H. G. & Gage, F. H. More hippocampal neurons in adult mice living in an enriched environment. Nature 386, 493–495 (1997).
Kempermann, G., Kuhn, H. G. & Gage, F. H. Experience-induced neurogenesis in the senescent dentate gyrus. J. Neurosci. 18, 3206–3212 (1998).
van Praag, H., Kempermann, G. & Gage, F. H. Running increases cell proliferation and neurogenesis in the adult mouse dentate gyrus. Nat. Neurosci. 2, 266–270 (1999).
Gonçalves, J. T., Schafer, S. T. & Gage, F. H. Adult neurogenesis in the hippocampus: from stem cells to behavior. Cell 167, 897–914 (2016).
Overstreet-Wadiche, L. S. & Westbrook, G. L. Functional maturation of adult-generated granule cells. Hippocampus 16, 208–215 (2006).
Götz, M., Nakafuku, M. & Petrik, D. Neurogenesis in the developing and adult brain-similarities and key differences. Cold Spring Harb. Perspect. Biol. 8, a018853 (2016).
Overstreet-Wadiche, L. S., Bensen, A. L. & Westbrook, G. L. Delayed development of adult-generated granule cells in dentate gyrus. J. Neurosci. 26, 2326–2334 (2006).
Shin, J. et al. Single-cell RNA-seq with waterfall reveals molecular cascades underlying adult neurogenesis. Cell Stem Cell 17, 360–372 (2015).
Habib, N. et al. Div-Seq: A single nucleus RNA-seq method reveals dynamics of rare adult newborn neurons in the CNS. Preprint at bioRxiv https://doi.org/10.1101/045989 (2016).
van Dongen, S. & Abreu-Goodger, C. in: Bacterial Molecular Networks. Methods in Molecular Biology (Methods and Protocols), (eds. van Helden, J., Toussaint, A. & Thieffry, D.) 281–295. (Springer, New York, 2012).
van der Maaten, L. & Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008).
Marques, S. et al. Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science 352, 1326–1329 (2016).
Hawrylycz, M. J. et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 489, 391–399 (2012).
Walker, T. L. et al. Lysophosphatidic acid receptor is a functional marker of adult hippocampal precursor cells. Stem Cell Rep. 6, 552–565 (2016).
Noctor, S. C., Martínez-Cerdeño, V. & Kriegstein, A. R. Distinct behaviors of neural stem and progenitor cells underlie cortical neurogenesis. J. Comp. Neurol. 508, 28–44 (2008).
Hsieh, J. Orchestrating transcriptional control of adult neurogenesis. Genes Dev. 26, 1010–1021 (2012).
Ponti, G. et al. Cell cycle and lineage progression of neural progenitors in the ventricular-subventricular zones of adult mice. Proc. Natl. Acad. Sci. USA 110, E1045–E1054 (2013).
Pastrana, E., Cheng, L.-C. & Doetsch, F. Simultaneous prospective purification of adult subventricular zone neural stem cells and their progeny. Proc. Natl. Acad. Sci. USA 106, 6387–6392 (2009).
Knobloch, M. et al. Metabolic control of adult neural stem cell activity by Fasn-dependent lipogenesis. Nature 493, 226–230 (2013).
Knobloch, M. et al. SPOT14-positive neural stem/progenitor cells in the hippocampus respond dynamically to neurogenic regulators. Stem Cell Rep. 3, 735–742 (2014).
Miller, J. A. et al. Conserved molecular signatures of neurogenesis in the hippocampal subgranular zone of rodents and primates. Development 140, 4633–4644 (2013).
Bonaguidi, M. A. et al. In vivo clonal analysis reveals self-renewing and multipotent adult neural stem cell characteristics. Cell 145, 1142–1155 (2011).
Nicola, Z., Fabel, K. & Kempermann, G. Development of the adult neurogenic niche in the hippocampus of mice. Front. Neuroanat. 9, 53 (2015).
Gilley, J. A., Yang, C.-P. & Kernie, S. G. Developmental profiling of postnatal dentate gyrus progenitors provides evidence for dynamic cell-autonomous regulation. Hippocampus 21, 33–47 (2011).
Enikolopov, G., Overstreet-Wadiche, L. & Ge, S. Viral and transgenic reporters and genetic analysis of adult neurogenesis. Cold Spring Harb. Perspect. Biol. 7, a018804 (2015).
Lawson, K. A. & Wilson, V. in Kaufman’s Atlas of Mouse Development Supplement (eds. Baldock, R., Bard, J., Davidson, D. R. & Morriss-Kay, G.) 51–64 (Academic Press, Oxford, 2016).
Kuckenberg, P., Kubaczka, C. & Schorle, H. The role of transcription factor Tcfap2c/TFAP2C in trophectoderm development. Reprod. Biomed. Online 25, 12–20 (2012).
Schemmer, J. et al. Transcription factor TFAP2C regulates major programs required for murine fetal germ cell maintenance and haploinsufficiency predisposes to teratomas in male mice. PLoS One 8, e71113 (2013).
Pinto, L. et al. AP2γ regulates basal progenitor fate in a region- and layer-specific manner in the developing cortex. Nat. Neurosci. 12, 1229–1237 (2009).
Mateus-Pinheiro, A. et al. AP2γ controls adult hippocampal neurogenesis and modulates cognitive, but not anxiety or depressive-like behavior. Mol. Psychiatry 22, 1725–1734 (2017).
Kim, W. et al. TFAP2C-mediated upregulation of TGFBR1 promotes lung tumorigenesis and epithelial-mesenchymal transition. Exp. Mol. Med. 48, e273 (2016).
Kang, J. et al. TFAP2C promotes lung tumorigenesis and aggressiveness through miR-183- and miR-33a-mediated cell cycle regulation. Oncogene 36, 1585–1596 (2017).
Zhuo, L. et al. Live astrocytes visualized by green fluorescent protein in transgenic mice. Dev. Biol. 187, 36–42 (1997).
Zeisel, A. et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 347, 1138–1142 (2015).
Acknowledgements
The authors thank P. Ernfors for critical discussion of the data and manuscript and A. Johnsson for lab management. The study was supported by the Knut and Alice Wallenberg foundation (2015.0041 to S.L.) and the Human Frontiers Science Program to A.Z. DNA sequencing was performed at the National Genomics Infrastructure, and single-cell RNA sequencing at the Eukaryotic Single-cell Genomics Facility, both at Science for Life Laboratory, Stockholm, Sweden.
Author information
Authors and Affiliations
Contributions
H.H., A.Z., and S.L. designed the study. H.H. and A.Z. performed all experiments, analyzed the data, and prepared the figures. P.L. analyzed the data. H.H. and S.L. wrote the paper, with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations
Integrated supplementary information
Supplementary Figure 1 Overview of sampling and technical details for Datasets A–C
(a) Schematic over the dentate gyrus granule layer emergence and maintained neurogenesis through development, as in Fig. 1a, with sampling timepoints indicated for each dataset. (b), (d), (f) Distribution of cell-type frequency in each dataset. Circle area represents fraction of positive cells, per sampling age, normalized by cell-type. Dashed box indicates time points sampled in the hGFAP:GFP reporter mouse, by FACS on GFP + cells. (c), (e), (g) Technical specifications of the datasets. Single-cell barplots for number of molecules (left) and genes (right), arranged by cluster. tSNE visualizations of each dataset (compare Fig. 1, Suppl. Figure 5, Fig. 5), are colored by number of genes (grey-low, red-high).
Supplementary Figure 2 Cajal–Retzius cells (CR) in the marginal zone of the dentate gyrus
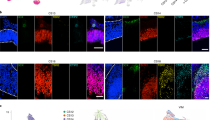
(a) t-SNE visualization of Dataset A (see Fig. 1). Cells are stained for expression of genes enriched in the Cajal-Retzius population (grey, low; red, high). (b) Allen Mouse Brain Atlas images of known Cajal-Retzius cell marker Reln (encoding reelin, co-expressed in hilus GABAergic neurons), and more specific genes identified in our dataset Lhx1 and Lhx5, across postnatal development and the adult. Scale bars represent 200μm. Image credit: Allen Institute. (c) Validation of Reln+/Lhx1+ Cajal-Retzius cells (outlined by solid circles) at ages P10, P16, P24 and P37, representative images from hybridizations on six sections in two mice per time point. Higher density of Cajal-Retzius cells along the marginal zone (dashed line) observed in younger ages. Co-expression of Reln/Lhx1 is preserved also in the adolescent. Left panel: Overview dentate gyrus, localization of Cajal-Retzius cells to the marginal zone. Middle panel: Zoom-in on area outlined by solid rectangle in left panel, spanning marginal zone (dashed line), molecular layer and granule layer (between solid outlines). Right panel: detailed view, zoom-in on area outlined by solid square in left panel. Scale bars represent 200μm (left panel) and 50μm (zoom-ins, middle and right panels). (d) Distribution of Cajal-Retzius cells in single-cell data from Dataset A (left) and B (right), across ages, shows enrichment in earlier postnatal time points.
Supplementary Figure 3 Early neurogenesis markers
t-SNE visualization of Dataset A (as in Fig. 1), cells are stained for expression of consecutive markers of early neurogenesis. Grey - low expression, red - high expression.
Supplementary Figure 4 Mutual KNN graph of early neurogenesis, without nIPC-specific genes
Analysis of astrocytes and early neurogenesis clusters (Radial Glia-like (n = 165), nIPC (n = 88), Neuroblast 1 (n = 97) and 2 (n = 777)) with nIPC-specific genes removed (see Methods, as in Fig. 2e-f). Nodes represent cells (total n = 1127 cells), edges connect mutual nearest neighbors. (a) Cells stained by cluster annotations as in Fig. 1d. nIPCs fall along a trajectory between Radial Glia-like and Neuroblast 1. (b) Mutual KNN graph stained by gene expression: progenitor genes (top), cell-cycle genes (middle), neurogenic genes (bottom) (grey, not expressed; red, expressed; compare Fig. 2f). Line indicates the divide of nIPCs by distance from Radial Glia-like or Neuroblast 1 (as in Fig. 2e).
Supplementary Figure 5 Time-resolved dentate gyrus Dataset B and characterization of hGFAP:GFP reporter mice
(a) t-SNE visualization of all 2303 cells, colored and labeled by cluster names. (b) Heatmap of 17 clusters from 2303 cells (all sampled time points), by their top marker genes (526 genes), expression normalized by gene. Barplots on top show sampling age source. (c) Dendrogram and correlation heatmap of clusters. (d) t-SNE visualization (as in (a)). Cells are colored by sampling age and experiment source (hGFAP:GFP sorted by FACS). Dashed box indicates astrocytes and clusters in early neurogenesis. (e) t-SNE visualization (as in (a)) with cells contributed by FACS are colored light green, cells expressing at least one molecule of Gfap or Cdk1 are encircled as indicated. Dashed circles outline populations that contain GFP+ cells, and are labeled by cluster name. (f) Analysis of clusters containing GFP+ cells. GFP expression is not restricted to Gfap+ cells or progenitor populations. Left: Contribution of GFP+ cells to each cluster, in percent. Line shows total contribution of GFP+ cells to the dataset (5.2%). Left center: Percentage of all cells expressing at least one molecule Gfap, per cluster. Right center: Percentage of GFP+ cells expressing at least one molecule Gfap, per cluster. Right: Percentage of all cells expressing at least one molecule Cdk1, per cluster. (g) Immunohistochemistry of hGFAP:GFP mouse dentate gyrus, stained with antibodies against GFP, GFAP, Aldoc (astrocytes) and Pdgfra (OPCs), representative images from stainings in two mice. Left panel: Overview of denate gyrus. Right panel: zoom-in, coexpression of respective markers indicated by arrowheads. Scale bars represent 200μm (left) and 50μm (zoom-ins, right).
Supplementary Figure 6 Markers of neurogenesis, maturation, and Notch-signaling
t-SNE visualization of Dataset A (as in Fig. 1d), cells stained for expression of markers of early neurogenesis (Calb2), granule cell maturation (Fxyd7-Icam5), and Notch-signaling related genes (Notch1, Notch2, Dner, Dlk2, Jag1). Grey, low expression; red, high expression.
Supplementary Figure 7 Overview of dentate gyrus Dataset C, spanning perinatal, juvenile, and adult neurogenesis
(a) t-SNE visualization of all 24,185 cells, colored and labeled by cluster names, sampled from perinatal (E16.5-P5), juvenile (P18-23) and adult (P120-132) mice. (b) Dendrogram and correlation heatmap of Dataset C clusters. (c) Heatmap of 24 clusters from 24,185 cells (all sampled time points), represented by their top marker genes (756 genes), expression normalized by gene. Barplots on top show sampling age source.
Supplementary Figure 8 Suggested model of cell types involved in perinatal and adult dentate neurogenesis
Both in perinatal and adult neurogenesis we propose distinct initiation clusters (radial glia and radial glia-like; size of circle symbolizes frequency), and convergence of the maturation process.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8
Supplementary Table 1
Gene enrichment in clusters of early neurogenesis
Supplementary Table 2
Early neurogenesis clusters: Pairwise Wilcoxon rank-sum test, q-values
Supplementary Table 3
Gene enrichment in clusters of granule cell maturation
Supplementary Table 4
Granule cell maturation clusters: Pairwise Wilcoxon rank-sum test, q-values
Supplementary Table 5
Experimental details of Datasets A and C sampling
Supplementary Table 6
Experimental details of Dataset B sampling
Rights and permissions
About this article
Cite this article
Hochgerner, H., Zeisel, A., Lönnerberg, P. et al. Conserved properties of dentate gyrus neurogenesis across postnatal development revealed by single-cell RNA sequencing. Nat Neurosci 21, 290–299 (2018). https://doi.org/10.1038/s41593-017-0056-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41593-017-0056-2
This article is cited by
-
DeepVelo: deep learning extends RNA velocity to multi-lineage systems with cell-specific kinetics
Genome Biology (2024)
-
Interrogations of single-cell RNA splicing landscapes with SCASL define new cell identities with physiological relevance
Nature Communications (2024)
-
Genetic labeling of embryonically-born dentate granule neurons in young mice using the PenkCre mouse line
Scientific Reports (2024)
-
The lncRNA Snhg11, a new candidate contributing to neurogenesis, plasticity, and memory deficits in Down syndrome
Molecular Psychiatry (2024)
-
Single-cell long-read sequencing-based mapping reveals specialized splicing patterns in developing and adult mouse and human brain
Nature Neuroscience (2024)