Abstract
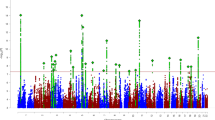
Delay discounting (DD), the tendency to discount the value of delayed versus current rewards, is elevated in a constellation of diseases and behavioral conditions. We performed a genome-wide association study of DD using 23,127 research participants of European ancestry. The most significantly associated single-nucleotide polymorphism was rs6528024 (P = 2.40 × 10−8), which is located in an intron of the gene GPM6B. We also showed that 12% of the variance in DD was accounted for by genotype and that the genetic signature of DD overlapped with attention-deficit/hyperactivity disorder, schizophrenia, major depression, smoking, personality, cognition and body weight.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Change history
08 January 2019
The author list was in the wrong order in the HTML version of the original article and in the HTML version of the original correction notice. This has been corrected to show the 23andMe Research Team as the fourth author and Abraham A. Palmer as the last author in both places.
11 May 2018
In the version of this article initially published, the consortium authorship was not presented correctly. The 23andMe Research Team was listed as the last author, rather than the fourth, and a line directing readers to the Supplementary Note for a list of members did appear but was not directly associated with the consortium name. Also, the Supplementary Note description stated that both member names and affiliations were included; in fact, only names are given. Finally, the URL for S-PrediXcan was given in the Methods as https://github.com/hakyimlab/S-PrediXcan; the correct URL is https://github.com/hakyimlab/MetaXcan. The errors have been corrected in the HTML and PDF versions of the article.
References
Bari, A. & Robbins, T. W. Prog. Neurobiol. 108, 44–79 (2013).
Hamilton, K. R. et al. Personal. Disord. 6, 182–198 (2015).
Jackson, J. N. S. & MacKillop, J. Biol. Psychiatry Cogn. Neurosci. Neuroimaging 1, 316–325 (2016).
Amlung, M., Vedelago, L., Acker, J., Balodis, I. & MacKillop, J. Addiction 112, 51–62 (2017).
McClelland, J. et al. Neurosci. Biobehav. Rev. 71, 506–528 (2016).
Insel, T. et al. Am. J. Psychiatry 167, 748–751 (2010).
Kirby, K. N., Petry, N. M. & Bickel, W. K. J. Exp. Psychol. Gen. 128, 78–87 (1999).
Anokhin, A. P., Grant, J. D., Mulligan, R. C. & Heath, A. C. Biol. Psychiatry 77, 887–894 (2015).
Yang, J., Lee, S. H., Goddard, M. E. & Visscher, P. M. Am. J. Hum. Genet. 88, 76–82 (2011).
Fjorback, A. W., Müller, H. K. & Wiborg, O. J. Mol. Neurosci. 37, 191–200 (2009).
Dere, E. et al. Behav. Brain Res. 277, 254–263 (2015).
Darna, M. et al. Behav. Brain Res. 293, 134–142 (2015).
Dalley, J. W. & Roiser, J. P. Neuroscience 215, 42–58 (2012).
Schweighofer, N. et al. J. Neurosci. 28, 4528–4532 (2008).
Fuchsova, B., Alvarez Juliá, A., Rizavi, H. S., Frasch, A. C. & Pandey, G. N. Neuroscience 299, 1–17 (2015).
MacKillop, J. J. Exp. Anal. Behav. 99, 14–31 (2013).
Gamazon, E. R. et al. Nat. Genet. 47, 1091–1098 (2015).
Bulik-Sullivan, B. et al. Nat. Genet. 47, 1236–1241 (2015).
Patros, C. H. G. et al. Clin. Psychol. Rev. 43, 162–174 (2016).
Gottesman, I. I. & Gould, T. D. Am. J. Psychiatry 160, 636–645 (2003).
Durand, E. Y., Do, C. B., Mountain, J. L. & Macpherson, J. M. Preprint at bioRxi v https://doi.org/10.1101/010512 (2014).
Eriksson, N. et al. PLoS. Genet. 6, e1000993 (2010).
Hyde, C. L. et al. Nat. Genet. 48, 1031–1036 (2016).
Lo, M.-T. et al. Nat. Genet. 49, 152–156 (2017).
Henn, B. M. et al. PLoS ONE 7, e34267 (2012).
Browning, S. R. & Browning, B. L. Am. J. Hum. Genet. 81, 1084–1097 (2007).
Fuchsberger, C., Abecasis, G. R. & Hinds, D. A. Bioinformatics 31, 782–784 (2015).
Zheng, X. et al. Pharmacogenomics J. 14, 192–200 (2014).
Gray, J. C., Amlung, M. T., Palmer, A. A. & MacKillop, J. J. Exp. Anal. Behav. 106, 156–163 (2016).
Saunders, J. B., Aasland, O. G., Babor, T. F., de la Fuente, J. R. & Grant, M. Addiction 88, 791–804 (1993).
Aschard, H., Vilhjálmsson, B. J., Joshi, A. D., Price, A. L. & Kraft, P. Am. J. Hum. Genet. 96, 329–339 (2015).
The 1000 Genomes Project Consortium. Nature 467, 1061–1073 (2010).
Willer, C. J., Li, Y. & Abecasis, G. R. Bioinformatics 26, 2190–2191 (2010).
Yang, J., Lee, S. H., Goddard, M. E. & Visscher, P. M. Methods. Mol. Biol. 1019, 215–236 (2013).
Bulik-Sullivan, B. K. et al. Nat. Genet. 47, 291–295 (2015).
International HapMap 3 Consortium. et al. Nature 467, 52–58 (2010).
Price, A. L., Zaitlen, N. A., Reich, D. & Patterson, N. Nat. Rev. Genet. 11, 459–463 (2010).
Benjamini, Y. & Hochberg, Y. J.R. Stat. Soc. 57, 289–300 (1995).
Sanchez-Roige, S. et al. Preprint at bioRxiv https://doi.org/10.1101/147397 (2017).
Acknowledgements
We thank the research participants and employees of 23andMe for making this work possible. J.M. was partially supported by the Peter Boris Chair in Addictions Research. S.S.-R. was supported by the Frontiers of Innovation Scholars Program (FISP; #3-P3029), the Interdisciplinary Research Fellowship in NeuroAIDS (IRFN; MH081482) and a pilot award from DA037844.
Author information
Authors and Affiliations
Consortia
Contributions
Conceptualization: A.A.P., J.M.; analysis and software: S.S.-R., P.F., L.K.D., J.C.G., A.A.P.; writing: S.S.-R., A.A.P.; review and editing: all authors.
Corresponding author
Ethics declarations
Competing interests
P.F., S.L.E., and members of the 23andMe Research Team are employees of 23andMe Inc. The opinions and assertions expressed herein are those of the authors; specifically, with respect to J.C.G., they do not reflect the official policy or position of the Uniformed Services University or the Department of Defense.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated Supplementary Information
Supplementary Figure 1 Regional association plot focusing on top SNP rs6528025 at 3′ of GPM6B gene on chromosome X at position 13.9 Mb
This plot was generated using LocusZoom1. The -log10(P value) is shown on the left y-axis; position in Mb is on the x-axis. Recombination rates (expressed in centiMorgans cM per Mb; NCBI Build GRCh37; highlighted in blue) are shown on the right y-axis. Pairwise linkage disequilibrium (r2) of each SNP with the top SNP in the region is indicated by its color. Crossed points represent imputed SNPs, circles represent directly genotyped SNPs. The statistical tests used were two-sided; sample size = 23,217.
Supplementary Figure 2 Regional association plot showing the second index SNP rs2665993, located in the EVPL gene on chromosome 17
This plot was generated using LocusZoom1. The -log10(P value) is shown on the left y-axis; position in Mb is on the x-axis. Recombination rates (expressed in centiMorgans cM per Mb; NCBI Build GRCh37; highlighted in blue) are shown on the right y-axis. Pairwise linkage disequilibrium (r2) of each SNP with the top SNP in the region is indicated by its color. Crossed points represent imputed SNPs, circles represent directly genotyped SNPs. The statistical tests used were two-sided; sample size = 23,217.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1 and 2
Supplementary Note
Consortia members & affiliations
Supplementary Data Set
Top 10,000 SNPs
Rights and permissions
About this article
Cite this article
Sanchez-Roige, S., Fontanillas, P., Elson, S.L. et al. Genome-wide association study of delay discounting in 23,217 adult research participants of European ancestry. Nat Neurosci 21, 16–18 (2018). https://doi.org/10.1038/s41593-017-0032-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41593-017-0032-x
This article is cited by
-
CRISPR/Cas9-mediated deletion of a GA-repeat in human GPM6B leads to disruption of neural cell differentiation from NT2 cells
Scientific Reports (2024)
-
Reward maximization assessed using a sequential patch depletion task in a large sample of heterogeneous stock rats
Scientific Reports (2023)
-
Relationship Between Parental Big Five And Children’s Ability To Delay Gratification
Journal of Child and Family Studies (2023)
-
Genetic Influences on Alcohol Sensitivity: a Critical Review
Current Addiction Reports (2023)
-
CADM2 is implicated in impulsive personality and numerous other traits by genome- and phenome-wide association studies in humans and mice
Translational Psychiatry (2023)