Abstract
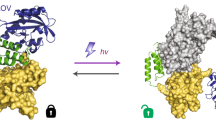
Lifeact is a popular peptide-based label of actin filaments in live cells. We have designed an improved Lifeact variant, LILAC, that binds to actin in light using the LOV2 protein. Light control allows the user to modulate actin labeling, enabling image analysis that leverages modulation for an enhanced view of F-actin dynamics in cells. Furthermore, the tool reduces actin perturbations and cell sickness caused by Lifeact overexpression.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
The data generated in this study are available as Source Data. The sequence of pMT-mCherry-LILAC has been deposited to GenBank (OP764687). Source data are provided with this paper.
References
Blanchoin, L., Boujemaa-Paterski, R., Sykes, C. & Plastino, J. Actin dynamics, architecture, and mechanics in cell motility. Physiol. Rev. 94, 235–263 (2014).
Melak, M., Plessner, M. & Grosse, R. Actin visualization at a glance. J. Cell Sci. 130, 525–530 (2017).
Riedl, J. et al. Lifeact: a versatile marker to visualize F-actin. Nat. Methods 5, 605–607 (2008).
Spracklen, A. J., Fagan, T. N., Lovander, K. E. & Tootle, T. L. The pros and cons of common actin labeling tools for visualizing actin dynamics during Drosophila oogenesis. Dev. Biol. 393, 209–226 (2014).
Flores, L. R., Keeling, M. C., Zhang, X., Sliogeryte, K. & Gavara, N. Author correction: Lifeact-TagGFP2 alters F-actin organization, cellular morphology and biophysical behaviour. Sci. Rep. 9, 9507 (2019).
Courtemanche, N., Pollard, T. D. & Chen, Q. Avoiding artefacts when counting polymerized actin in live cells with LifeAct fused to fluorescent proteins. Nat. Cell Biol. 18, 676–683 (2016).
Belyy, A., Merino, F., Sitsel, O. & Raunser, S. Structure of the Lifeact–F-actin complex. PLoS Biol. 18, e3000925 (2020).
Kumari, A., Kesarwani, S., Javoor, M. G., Vinothkumar, K. R. & Sirajuddin, M. Structural insights into actin filament recognition by commonly used cellular actin markers. EMBO J. 39, e104006 (2020).
Crosson, S. & Moffat, K. Structure of a flavin-binding plant photoreceptor domain: insights into light-mediated signal transduction. Proc. Natl Acad. Sci. USA 98, 2995–3000 (2001).
Kennis, J. T. M. et al. The LOV2 domain of phototropin: a reversible photochromic switch. J. Am. Chem. Soc. 126, 4512–4513 (2004).
Halavaty, A. S. & Moffat, K. N- and C-terminal flanking regions modulate light-induced signal transduction in the LOV2 domain of the blue light sensor phototropin 1 from Avena sativa. Biochemistry 46, 14001–14009 (2007).
Strickland, D. et al. TULIPs: tunable, light-controlled interacting protein tags for cell biology. Nat. Methods 9, 379–384 (2012).
French, A. R., Sosnick, T. R. & Rock, R. S. Investigations of human myosin VI targeting using optogenetically controlled cargo loading. Proc. Natl Acad. Sci. USA 114, E1607–E1616 (2017).
Niopek, D. et al. Engineering light-inducible nuclear localization signals for precise spatiotemporal control of protein dynamics in living cells. Nat. Commun. 5, 4404 (2014).
Guntas, G. et al. Engineering an improved light-induced dimer (iLID) for controlling the localization and activity of signaling proteins. Proc. Natl Acad. Sci. USA 112, 112–117 (2015).
Strickland, D., Moffat, K. & Sosnick, T. R. Light-activated DNA binding in a designed allosteric protein. Proc. Natl Acad. Sci. USA 105, 10709–10714 (2008).
Zayner, J. P., Antoniou, C. & Sosnick, T. R. The amino-terminal helix modulates light-activated conformational changes in AsLOV2. J. Mol. Biol. 419, 61–74 (2012).
Zayner, J. P., Antoniou, C., French, A. R., Hause, R. J. & Sosnick, T. R. Investigating models of protein function and allostery with a widespread mutational analysis of a light-activated protein. Biophys. J. 105, 1027–1036 (2013).
Zayner, J. P., Mathes, T., Sosnick, T. R. & Kennis, J. T. M. Helical contributions mediate light-activated conformational change in the LOV2 domain of Avena sativa phototropin 1. ACS Omega 4, 1238–1243 (2019).
Zayner, J. P. & Sosnick, T. R. Factors that control the chemistry of the LOV domain photocycle. PLoS One 9, e87074 (2014).
Alexandre, M. T. A., Arents, J. C., van Grondelle, R., Hellingwerf, K. J. & Kennis, J. T. M. A base-catalyzed mechanism for dark state recovery in the Avena sativa phototropin-1 LOV2 domain. Biochemistry 46, 3129–3137 (2007).
Harper, S. M., Neil, L. C. & Gardner, K. H. Structural basis of a phototropin light switch. Science 301, 1541–1544 (2003).
Matsuoka, D. & Tokutomi, S. Blue light-regulated molecular switch of Ser/Thr kinase in phototropin. Proc. Natl Acad. Sci. USA 102, 13337–13342 (2005).
Harper, S. M., Christie, J. M. & Gardner, K. H. Disruption of the LOV−Jα helix interaction activates phototropin kinase activity. Biochemistry 43, 16184–16192 (2004).
Rogers, S. L., Wiedemann, U., Stuurman, N. & Vale, R. D. Molecular requirements for actin-based lamella formation in Drosophila S2 cells. J. Cell Biol. 162, 1079–1088 (2003).
Marriott, G. et al. Optical lock-in detection imaging microscopy for contrast-enhanced imaging in living cells. Proc. Natl Acad. Sci. USA 105, 17789–17794 (2008).
Strickland, D. et al. Rationally improving LOV domain-based photoswitches. Nat. Methods 7, 623–626 (2010).
Han, K. An efficient DDAB-mediated transfection of Drosophila S2 cells. Nucleic Acids Res. 24, 4362–4363 (1996).
Buster, D. W., Nye, J., Klebba, J. E. & Rogers, G. C. Preparation of Drosophila S2 cells for light microscopy. J. Vis. Exp. 40, 1982 (2010).
Ruijgrok, P. V. et al. Optical control of fast and processive engineered myosins in vitro and in living cells. Nat. Chem. Biol. 17, 540–548 (2021).
Heier, J. A., Dickinson, D. J. & Kwiatkowski, A. V. Measuring protein binding to F-actin by co-sedimentation. J. Vis. Exp. 123, 55613 (2017).
Acknowledgements
This work was supported by grant numbers EB009412, GM055694 (T.R.S) and GM124272 (R.S.R) awarded by the National Institutes of Health, as well as the David Grier Prize for Innovative Research in Biophysical Sciences and the 2022 American Heart Association Predoctoral Fellowship 915957 (K.L.K).
Author information
Authors and Affiliations
Contributions
Author contributions: K.L.K., A.R.F., T.R.S., and R.S.R. designed research; K.L.K. performed research; K.L.K., T.R.S., and R.S.R. contributed new reagents/analytic tools; K.L.K., T.R.S., and R.S.R. analyzed data; and K.L.K., A.R.F., T.R.S., and R.S.R. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
K.L.K., A.R.F., T.R.S. and R.S.R. have submitted a patent application (PCT/US2022/077754; Methods and Compositions Comprising Actin Binding Proteins) to the USA patent office.
Peer review
Peer review information
Nature Methods thanks Michael Sixt, Roland Wedlich-Söldner and the other, anonymous, reviewer for their contribution to the peer review of this work. Peer reviewer reports are available. Primary Handling Editor: Nina Vogt, in collaboration with the Nature Methods team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
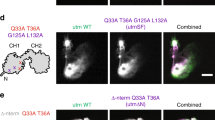
Extended Data Fig. 1 LILAC highlights actin structures through background subtraction.
TIRF image of an S2 cell expressing LILAC without (a) and with (b) pre-excitation image background subtraction to eliminate cytosolic background. Kymographs of the region in yellow without (c) and with (d) pre-excitation background subtraction. (e) Histograms of normalized pixel intensities in the outer ring of the cell with (CV = 0.58) and without (CV = 0.23) image subtraction.
Extended Data Fig. 2 LILAC labels actin patches in S2 cells.
TIRF images of live S2 cells expressing mCherry-LILAC taken pre- and post-488 nm (blue) laser excitation. Scale bar is 5 μm. Inset images are 3x magnified.
Extended Data Fig. 3 LILAC labels filopodia in landing S2 cells.
TIRF images of live S2 cells expressing mCherry-LILAC taken pre- and post-488 nm (blue) laser excitation. Cells are imaged as they land on the ConA coated coverslip. Scale bar is 5 μm. Inset images are 2x magnified.
Extended Data Fig. 4 Phalloidin, Lifeact, and LILAC co-label actin structures in S2 cells.
TIRF images of fixed S2 cells expressing (a) mCherry-Lifeact or (b) mCherry-LILAC stained with phalloidin. Cells were fixed under blue light. Pearson’s correlation coefficients are displayed on the merge image. Scale bar is 5 μm.
Extended Data Fig. 5 Image processing methods with LILAC.
(a) Post-excitation image of an S2 cell with pre-excitation image subtracted. (b) OLID (optical lock-in detection)26 image of an S2 cell. White pixels are correlated, black anticorrelated, and mid-gray are uncorrelated. We used the first 50 frames of a movie (0.5 s/frame) where the excitation pulse arrives in frame 2 to generate the OLID image. An S2 cell expressing mCherry-LILAC before (c) and after (d) blue light excitation. The outer lamellipodial ring and interior disk of cells are hand traced in FIJI (yellow). Scale bar, 5 µm. Actin labeling ratio: ALR = (mean pixel intensity)outer/(mean pixel intensity)interior, actin switching ratio: ASR = ALRpost/ALRpre.
Extended Data Fig. 6 Quantification of labeling and switching of LILAC.
Max actin labeling ratio (a, R2 = 0.27, P = 2.3*10−6, n = 72) and switching ratio (b, R2 = 0.13, P = 1.3*10−3, n = 72) of LILAC-expressing cells as a function of cell intensity. Maximum actin labeling (c, P = 0.11) and switching ratios (d, P = 9.66*10−15) of LILAC, Lifeact, and the light-mimic mutant, LILACI539E. The center line represents the median. Boxes span the 25th and 75th percentiles. Whiskers extend from the 25th percentile − 1.5x the interquartile range (IQR) and from the 75th percentile + 1.5x IQR. Data beyond the whiskers represent outliers and are plotted individually. For c and d, P values were determined initially using a one-way ANOVA test (n = 32 for Lifeact, n = 18 for LILAC, n = 23 for LILACI539E). A post-hoc Dunn test was used to determine pairwise P values (LILAC-Lifeact: P = 1.7*10−15, LILAC-LILACI593E: P = 1.4*10−5, Lifeact-LILACI593E: P = 2.4*10−3) (**= P ≤ 0.01, ***= P ≤ 0.001, ****= P ≤ 0.0001).
Extended Data Fig. 7 Binding of Lifeact and LILAC to F-actin.
a,b, SDS-PAGE of the protein of interest (P), either mCherry- LILACI539E (a) or mCherry-Lifeact (b), in the pellet after co-sedimentation with 0.5 μM actin (A). Concentration of free protein added to each lane is labeled above in μM. c, Free protein plotted against bound protein in the pellet after co-sedimentation to determined binding affinities of Lifeact (Kd=3.9 μM) and LILAC539E (Kd=3.4 μM). Data were normalized to the maximum observed and fitted to a hyperbola. The constitutively active form of LILAC was used because excitation of LILAC cannot be maintained in the dark ultracentrifuge.
Extended Data Fig. 8 LILAC excitation dynamics.
a, Min-max normalized actin labeling ratio traces for cells initially in the dark, then activated with blue light continually through the end of the movie. b, Max switching ratios for cells excited for various lengths of time at 1% laser power, colored by cell.
Extended Data Fig. 9 Dark state recovery time constants are cell specific and tunable.
a, First and second recovery time constants for individual cells expressing wild type (R2 = 0.70, n = 34) and T406A/T407A (R2 = 0.56, n = 26) LILAC. Gray line is X = Y. b, Kernel density estimations of the first time constants for wild type (blue) and T406A/T407A (lilac) LILAC. Mean is indicated by vertical lines. c, Min-max normalized actin labeling ratio traces of LILAC(T406A/T407A)expressing cells with (teal) and without (lilac) 1 mM imidazole. Cells were activated with blue light every 60 and 130 seconds respectively. d, First and second recovery time constants for individual cells expressing LILAC(T406A/T407A) with varying imidazole concentrations (R2 = 0.77, n = 128, Pearson P = 7.3e-46). Gray line is X = Y. e, First recovery time constants as a function of imidazole concentration (R2 = 0.52, Pearson P = 3.5e-24) with a linear regression fit to log-transformed data and shaded regions indicating the 95% confidence interval. Data with 0 and 1 mM imidazole were taken at different times with different starter cultures of S2 cells than all other concentrations, which were imaged on the same day.
Extended Data Fig. 10 LILAC constructs.
Schematics of LILAC (a) and other vectors used. a, Schematic of LILAC, including LOV2opt(AsLOV2401-543 G528A/I532A/N538E). Vectors used for protein expression (b,c) and expression in S2 cells (d–f), where TEV is the TEV protease cleavage site.
Supplementary information
Supplementary Video 1
Light-induced labeling of lamellipodia with LILAC. TIRF imaging of an S2 cell expressing mCherry-tagged LILAC. Blue light (488 nm) is pulsed for 0.5 s after the first frame, and again at 1 min.
Supplementary Video 2
Labeling with LILAC enables enhanced imaging with pre-excitation subtraction. TIRF imaging of an S2 cell expressing mCherry-tagged LILAC, with the pre-excitation image subtracted from all images in the movie. Negative values are all floored to 0 (black). Blue light (488 nm) is pulsed for 0.5 s after the first frame, and again at 1 min.
Supplementary Video 3
Sustained light-induced labeling of lamellipodia with LILAC. TIRF imaging of an S2 cell expressing mCherry-tagged LILAC. Blue light (488 nm) is continually exciting the cell after the first frame.
Supplementary Video 4
Labeling with LILAC enables enhanced imaging with pre-excitation subtraction. TIRF imaging of an S2 cell expressing mCherry-tagged LILAC, with the pre-excitation image subtracted from all images in the movie. Negative values are all floored to 0 (black). Blue light (488 nm) is continually exciting the cell after the first frame.
Source data
Source Data Fig. 1
Statistical Source Data
Source Data Fig. 2
Statistical Source Data
Source Data Extended Data Fig. 1
Statistical Source Data
Source Data Extended Data Fig. 6
Statistical Source Data
Source Data Extended Data Fig. 7
Unprocessed gel of mCherry-LILACI539E + F-actin co-sedimentation. Lane 1 is the ladder, lanes 2–8 are pre-spin and lanes 9–15 are post-spin. Unprocessed gel of mCherry-Lifeact + F-actin co-sedimentation. Lane 1 is the ladder, lanes 2–8 are pre-spin and lanes 9–15 are post-spin. Statistical Source Data
Source Data Extended Data Fig. 8
Statistical Source Data
Source Data Extended Data Fig. 9
Statistical Source Data
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kroll, K.L., French, A.R., Sosnick, T.R. et al. LILAC: enhanced actin imaging with an optogenetic Lifeact. Nat Methods 20, 214–217 (2023). https://doi.org/10.1038/s41592-022-01761-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-022-01761-3