Abstract
In situ cryo electron tomography of cryo focused ion beam milled samples has emerged in recent years as a powerful technique for structural studies of macromolecular complexes in their native cellular environment. However, the possibilities for recording tomographic tilt series in a high-throughput manner are limited, in part by the lamella-shaped samples. Here we utilize a geometrical sample model and optical image shift to record tens of tilt series in parallel, thereby saving time and gaining access to sample areas conventionally used for tracking specimen movement. The parallel cryo electron tomography (PACE-tomo) method achieves a throughput faster than 5 min per tilt series and allows for the collection of sample areas that were previously unreachable, thus maximizing the amount of data from each lamella. Performance testing with ribosomes in vitro and in situ on state-of-the-art and general-purpose microscopes demonstrated the high throughput and quality of PACE-tomo.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Subtomogram averages were deposited in the Electron Microscopy Data Bank under accession codes EMD-33834, EMD-33115, EMD-33116, EMD-33117, EMD-33118 and EMD-33833. Raw tilt series frames were deposited in the Electron Microscopy Public Image Archive under accession codes EMPIAR-11111, EMPIAR-10985, EMPIAR-10986 and EMPIAR-10987.
Code availability
SerialEM python scripts for auxiliary functions, target selection and PACE-tomo are available at https://github.com/eisfabian/PACEtomo.
References
Hylton, R. K. & Swulius, M. T. Challenges and triumphs in cryo-electron tomography. iScience 24, 102959 (2021).
Assaiya, A., Burada, A. P., Dhingra, S. & Kumar, J. An overview of the recent advances in cryo-electron microscopy for life sciences. Emerg. Top. Life Sci. 5, 151–168 (2021).
Danev, R. et al. Routine sub-2.5 Å cryo-EM structure determination of GPCRs. Nat. Commun. 12, 4333 (2021).
Hagen, W. J. H., Wan, W. & Briggs, J. A. G. Implementation of a cryo-electron tomography tilt-scheme optimized for high resolution subtomogram averaging. J. Struct. Biol. 197, 191–198 (2017).
Schur, F. K. M. et al. An atomic model of HIV-1 capsid-SP1 reveals structures regulating assembly and maturation. Science 353, 506–508 (2016).
Tegunov, D., Xue, L., Dienemann, C., Cramer, P. & Mahamid, J. Multi-particle cryo-EM refinement with M visualizes ribosome-antibiotic complex at 3.5 Å in cells. Nat. Methods 18, 186–193 (2021).
Chreifi, G., Chen, S., Metskas, L. A., Kaplan, M. & Jensen, G. J. Rapid tilt-series acquisition for electron cryotomography. J. Struct. Biol. 205, 163–169 (2019).
Eisenstein, F., Danev, R. & Pilhofer, M. Improved applicability and robustness of fast cryo-electron tomography data acquisition. J. Struct. Biol. 208, 107–114 (2019).
Cheng, A. et al. High resolution single particle cryo-electron microscopy using beam-image shift. J. Struct. Biol. 204, 270–275 (2018).
Weis, F. & Hagen, W. J. H. Combining high throughput and high quality for cryo-electron microscopy data collection. Acta Crystallogr. D Struct. Biol. 76, 724–728 (2020).
Wu, C., Huang, X., Cheng, J., Zhu, D. & Zhang, X. High-quality, high-throughput cryo-electron microscopy data collection via beam tilt and astigmatism-free beam-image shift. J. Struct. Biol. 208, 107396 (2019).
Danev, R., Yanagisawa, H. & Kikkawa, M. Cryo-EM performance testing of hardware and data acquisition strategies. Microscopy 70, 487–497 (2021).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018).
Bouvette, J. et al. Beam image-shift accelerated data acquisition for near-atomic resolution single-particle cryo-electron tomography. Nat. Commun. 12, 1957 (2021).
Zheng, Q. S., Braunfeld, M. B., Sedat, J. W. & Agard, D. A. An improved strategy for automated electron microscopic tomography. J. Struct. Biol. 147, 91–101 (2004).
Albert, S. et al. Direct visualization of degradation microcompartments at the ER membrane. Proc. Natl Acad. Sci. USA 117, 1069–1080 (2020).
Gupta, T. K. et al. Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity. Cell 184, 3643–3659 (2021).
Mahamid, J. et al. Visualizing the molecular sociology at the HeLa cell nuclear periphery. Science 351, 969–972 (2016).
Schaffer, M. et al. A cryo-FIB lift-out technique enables molecular-resolution cryo-ET within native Caenorhabditis elegans tissue. Nat. Methods 16, 757–762 (2019).
Weiss, G. L. et al. Structure of a thylakoid-anchored contractile injection system in multicellular cyanobacteria. Nat. Microbiol. 7, 386–396 (2022).
Marko, M., Hsieh, C., Schalek, R., Frank, J. & Mannella, C. Focused-ion-beam thinning of frozen-hydrated biological specimens for cryo-electron microscopy. Nat. Methods 4, 215–217 (2007).
Medeiros, J. M. et al. Robust workflow and instrumentation for cryo-focused ion beam milling of samples for electron cryotomography. Ultramicroscopy 190, 1–11 (2018).
Schaffer, M. et al. Optimized cryo-focused ion beam sample preparation aimed at in situ structural studies of membrane proteins. J. Struct. Biol. 197, 73–82 (2017).
Buckley, G. et al. Automated cryo-lamella preparation for high-throughput in-situ structural biology. J. Struct. Biol. 210, 107488 (2020).
Klumpe, S. et al. A modular platform for automated cryo-FIB workflows. eLife 10, e70506 (2021).
Tacke, S. et al. A streamlined workflow for automated cryo focused ion beam milling. J. Struct. Biol. 213, 107743 (2021).
Zachs, T. et al. Fully automated, sequential focused ion beam milling for cryo-electron tomography. eLife 9, e52286 (2020).
Khavnekar, S. et al. Multishot tomography for high-resolution in situ subtomogram averaging. Preprint at bioRxiv https://doi.org/10.1101/2022.04.10.487763 (2022).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Schorb, M., Haberbosch, I., Hagen, W. J. H., Schwab, Y. & Mastronarde, D. N. Software tools for automated transmission electron microscopy. Nat. Methods 16, 471 (2019).
Harris, C. R. et al. Array programming with NumPy. Nature 585, 357–362 (2020).
Virtanen, P. et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods 17, 261–272 (2020).
Khoshouei, M., Pfeffer, S., Baumeister, W., Förster, F. & Danev, R. Subtomogram analysis using the Volta phase plate. J. Struct. Biol. 197, 94–101 (2017).
Zivanov, J. et al. A Bayesian approach to single-particle electron cryo-tomography in RELION-4.0. Preprint at bioRxiv https://doi.org/10.1101/2022.02.28.482229 (2022).
Khavnekar, S. et al. Optimizing cryo-FIB lamellas for sub-5Å in situ structural biology. Preprint at bioRxiv https://doi.org/10.1101/2022.06.16.496417 (2022).
Wang, Z. et al. Structures from intact myofibrils reveal mechanism of thin filament regulation through nebulin. Science 375, eabn1934 (2022).
Peck, A. et al. Montage electron tomography of vitrified specimens. J. Struct. Biol. 214, 107860 (2022).
Yang, J. E. et al. Correlative cryogenic montage electron tomography for comprehensive in-situ whole-cell structural studies. Preprint at bioRxiv https://doi.org/10.1101/2021.12.31.474669 (2022).
Mastronarde, D. N. Correction for non-perpendicularity of beam and tilt axis in tomographic reconstructions with the IMOD package. J. Microsc. 230, 212–217 (2008).
Zheng, S. et al. AreTomo: an integrated software package for automated marker-free, motion-corrected cryo-electron tomographic alignment and reconstruction. J. Struct. Biol. X 6, 100068 (2022).
Ni, T. et al. High-resolution in situ structure determination by cryo-electron tomography and subtomogram averaging using emClarity. Nat. Protoc. 17, 421–444 (2022).
Chen, M. et al. A complete data processing workflow for cryo-ET and subtomogram averaging. Nat. Methods 16, 1161–1168 (2019).
Burt, A., Gaifas, L., Dendooven, T. & Gutsche, I. A flexible framework for multi-particle refinement in cryo-electron tomography. PLoS Biol. 19, e3001319 (2021).
Ecken, J., von der, Heissler, S. M., Pathan-Chhatbar, S., Manstein, D. J. & Raunser, S. Cryo-EM structure of a human cytoplasmic actomyosin complex at near-atomic resolution. Nature 534, 724–728 (2016).
Fialka, I. et al. The estrogen-dependent c-JunER protein causes a reversible loss of mammary epithelial cell polarity involving a destabilization of adherens junctions. J. Cell Biol. 132, 1115–1132 (1996).
Rigort, A. et al. Focused ion beam micromachining of eukaryotic cells for cryoelectron tomography. Proc. Natl Acad. Sci. USA 109, 4449–4454 (2012).
Wolff, G. et al. Mind the gap: micro-expansion joints drastically decrease the bending of FIB-milled cryo-lamellae. J. Struct. Biol. 208, 107389 (2019).
Ederth, J., Mandava, C. S., Dasgupta, S. & Sanyal, S. A single-step method for purification of active His-tagged ribosomes from a genetically engineered Escherichia coli. Nucleic Acids Res. 37, e15 (2009).
Kremer, J. R., Mastronarde, D. N. & McIntosh, J. R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76 (1996).
Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Zhang, K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Wagner, T. et al. SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun. Biol. 2, 1–13 (2019).
Castaño-Díez, D., Kudryashev, M., Arheit, M. & Stahlberg, H. Dynamo: a flexible, user-friendly development tool for subtomogram averaging of cryo-EM data in high-performance computing environments. J. Struct. Biol. 178, 139–151 (2012).
Rigort, A. et al. Automated segmentation of electron tomograms for a quantitative description of actin filament networks. J. Struct. Biol. 177, 135–144 (2012).
Heumann, J. M., Hoenger, A. & Mastronarde, D. N. Clustering and variance maps for cryo-electron tomography using wedge-masked differences. J. Struct. Biol. 175, 288–299 (2011).
Nicastro, D. et al. The molecular architecture of axonemes revealed by cryoelectron tomography. Science 313, 944–948 (2006).
Goddard, T. D. et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
Buchholz, T.-O. et al. Content-aware image restoration for electron microscopy. Methods Cell Biol. 152, 277–289 (2019).
Acknowledgements
We are grateful to Y. Fujiyoshi and H. Suzuki for stimulating discussions and valuable feedback. We thank K. Nakamura and Y. Sakamaki for the management and support of the Graduate School of Medicine cryoEM facility. We are grateful for support of cryoFIB milling and instrument maintenance by Y. Fukuda and for comments on the manuscript by I. Selvam. This research was partially supported by Platform Project for Supporting Drug Discovery and Life Science Research (Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS)) from AMED under grant number JP21am0101115j0005. F.E. is an International Research Fellow of the Japan Society for the Promotion of Science (JSPS, #P20764) and received a Grant-in-Aid for Scientific Research (KAKENHI, 21F20764). H.Y. and M.K. were supported by Grant-in-Aid for Transformative Research Areas A (JSPS, 21H05248). S.T. received funding from Grant-in-Aid for Specially Promoted Research (JP19H05468). R.D. was supported by Takeda Science Foundation 2019 Medical Research Grant and Japan Science and Technology Agency PRESTO (18069571).
Author information
Authors and Affiliations
Contributions
F.E. developed PACE-tomo and conducted all experiments. H.Y. purified ribosomes and processed several datasets. H.K. prepared the eukaryotic cell cultures. F.E., H.Y., H.K., M.K., S.T. and R.D. designed the experiments. F.E. wrote the manuscript with comments from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Methods thanks Petr Chlanda, Wim Hagen, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling editor: Rita Strack, in collaboration with the Nature Methods team. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 A linear defocus ramp can compensate for errors in tilt axis offset.
a. Shown are defocus values estimated by CTF fitting for two tilt series collected on the same lamella using PACE-tomo. One tilt series was recorded without applying a defocus ramp (dashed line). The other tilt series (solid line) was recorded while applying a defocus ramp of 0.03 µm/degree. Outliers at extreme tilt angles were not considered for the linear fit of the defocus slope. b. Shown are defocus values estimated by CTF fitting for two tilt series collected on amorphous carbon support using PACE-tomo. One tilt series was recorded using the tilt axis offset determined by the fine eucentricity routine of SerialEM (dashed line). The other tilt series (solid line) was recorded using the tilt axis offset determined using the PACEtomo_measureOffset.py script.
Extended Data Fig. 2 Side entry holder behavior throughout dose-symmetric tilt series.
a. Specimen shifts in µm along x and y (parallel and perpendicular to the tilt axis respectively). The behavior of the negative tilt angle branch differs from the positive tilt angle branch. b. Defocus throughout the tilt series measured by beam tilt. While defocus remains relatively constant for the positive tilt angle branch, it shows a significant slope for the negative tilt angle branch.
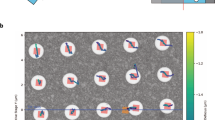
Extended Data Fig. 3 PACE-tomo tilt series collected using a cryo side-entry holder.
a. An overview image of a holey carbon sample area indicating the positions at which tilt series were collected. Targets are numbered according to acquisition order. Red frame indicates the position of the tracking tilt series. The tilt axis is along Y = 0. Collection time was 118 minutes for 25 tilt series with an angular range of ± 30°. b, c. Measured specimen shift errors by cross correlation-based alignment to first tilt image at 0° in each tilt series in x- (b) and y-direction (c), parallel and perpendicular to the tilt axis, respectively. Tilt series 1 is the tracking tilt series and points are coloured grey. d. Defocus values for each tilt series estimated by CTF fitting. e. Defocus values for each tilt series estimated by CTF fitting of the 0° tilt image plotted against the target coordinates in X and Y. The 3D plot shows that targets are approximately on a plane indicating significant tilt of the sample support film.
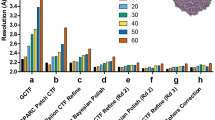
Extended Data Fig. 4 Image shift influence on resolution.
a. Tilt series of dataset 1 were divided into 5 groups parallel (red groups) and perpendicular (blue groups) to the tilt axis, respectively, and reconstructed independently. Resolutions were plotted against the relative distance to the central group. Neither parallel nor perpendicular groups to the tilt axis show a significant resolution dependency on the distance from the centre. b. Acquisition pattern for dataset 2 – PACE-tomo with 81 targets in a regular 9 by 9 pattern. Targets are numbered according to acquisition order. The tracking tilt series (red frame) is at the origin and all other targets are acquired by relative image shift along the tilt axis (x-axis) and perpendicular to the tilt axis (y-axis). Collection time was 6 hours. c. Vectors representing beam tilt determined by RELION Tomo CTF refinement using 1 optics group per tilt series. Origins of vectors are at their respective image shifts relative to the tracking tilt series at 0° tilt angle. Magnitude scale of vectors is indicated by the scale bar (top right). d. Tilt series of dataset 2 were divided into 5 groups parallel (red groups) and perpendicular (blue groups) to the tilt axis, respectively, and reconstructed independently. Resolutions were plotted against the relative distance to the central group. Parallel groups to the tilt axis show a resolution dependency on the distance from the centre while perpendicular groups do not.
Extended Data Fig. 5 PACE-tomo facilitates data collection in challenging areas.
a. Shown is a 0.66 nm thick slice through one of the 21 cryo electron tomograms (acquisition area 3) collected on the lamella shown in Fig. 4b. The subtomogram average was mapped into the tomogram using the refined coordinates and orientations. Membrane associated and cytoplasmic ribosomes were coloured red and blue, respectively. Scale bar: 100 nm. b. Shown is a 0.66 nm thick slice through one of 7 cryo electron tomograms collected on the bottom lamella shown in c. The subtomogram average of actin was mapped into the tomogram using the refined coordinates and orientations. Scale bar: 100 nm. c. Overview montages of two (out of 41) cryoFIB-milled lamellae indicating the positions at which tilt series were collected. Red frame indicates the position that was used for the tracking tilt series. White arrowheads mark targets that would not have been accessible by conventional cryoET collection schemes. Collection times were 23 minutes for 6 tilt series (top) and 30 minutes for 7 tilt series (bottom, used in dataset 5).
Extended Data Fig. 6 Representative sample used for cryoFIB milling.
a. Scanning electron microscopy image of a representative sample of EpH4 cells grown on a 150-mesh gold grid. A total of eight grids were used to prepare 41 lamellae. Scale bar: 1 mm. b. CryoFIB image after premilling trenches at 38° that mark targets for lamella milling and limit lamella length. Shown are four examples of the 41 prepared lamellae. Scale bar: 100 µm. c. Scanning electron microscopy image of finished lamella with micro-expansion joints that was used for a PACE-tomo acquisition (Extended Data Fig. 5c top). Shown is one of the 41 prepared lamellae. Scale bar: 10 µm.
Extended Data Fig. 7 Simplified schematic of PACE-tomo.
Shown are the essential steps of the PACE-tomo data acquisition scheme.
Extended Data Fig. 8 Processing workflows.
Shown are the processing workflows for datasets 1–5 using RELION-4.0-beta. Blue and red boxes indicate steps done in dynamo and PEET, respectively. Final maps were filtered and coloured according to the local resolution distribution.
Supplementary information
Supplementary Information
Supplementary Table 1, and captions for Supplementary Videos 1 and 2.
Supplementary Video 1
Slices through tomogram from dataset 4 collected on the cryoFIB-milled lamella shown in Fig. 4b (tilt series 3). The subtomogram average was mapped into the tomogram using the refined coordinates and orientations. Membrane-associated and cytoplasmic ribosomes were coloured red and blue, respectively.
Supplementary Video 2
Cross-correlation aligned tilt series from dataset 4 collected on the cryoFIB-milled lamella shown in Fig. 4b (tilt series 3). Scale bar, 100 nm.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Eisenstein, F., Yanagisawa, H., Kashihara, H. et al. Parallel cryo electron tomography on in situ lamellae. Nat Methods 20, 131–138 (2023). https://doi.org/10.1038/s41592-022-01690-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-022-01690-1
This article is cited by
-
Stepwise assembly and release of Tc toxins from Yersinia entomophaga
Nature Microbiology (2024)
-
Bridging structural and cell biology with cryo-electron microscopy
Nature (2024)
-
Square beams for optimal tiling in transmission electron microscopy
Nature Methods (2024)
-
Unlocking cryo-EM’s multishot potential with square or rectangular beams
Nature Methods (2024)
-
The application and development of electron microscopy for three-dimensional reconstruction in life science: a review
Cell and Tissue Research (2024)