Abstract
DNA–protein interactions mediate physiologic gene regulation and may be altered by DNA variants linked to polygenic disease. To enhance the speed and signal-to-noise ratio (SNR) in the identification and quantification of proteins associated with specific DNA sequences in living cells, we developed proximal biotinylation by episomal recruitment (PROBER). PROBER uses high-copy episomes to amplify SNR, and proximity proteomics (BioID) to identify the transcription factors and additional gene regulators associated with short DNA sequences of interest. PROBER quantified both constitutive and inducible association of transcription factors and corresponding chromatin regulators to target DNA sequences and binding quantitative trait loci due to single-nucleotide variants. PROBER identified alterations in regulator associations due to cancer hotspot mutations in the hTERT promoter, indicating that these mutations increase promoter association with specific gene activators. PROBER provides an approach to rapidly identify proteins associated with specific DNA sequences and their variants in living cells.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The mass spectrometry proteomics raw data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifiers PXD023732 (YY1, NF-κB, SNP rs7296179 and hTERT promoter PROBER-MS), PXD029470 (YY1 endogenous loci and U2OS cell line PROBER-MS, DNA pull-down and BioID mass spectrometry) and PXD032726 (PROBER-MS using on-bead sample processing). Raw data for STAT1 PROBER-MS are not available, therefore spectral search files (PSM) were deposited in PXD023732. Supplementary Table 29 lists the data deposited. Figure panels that were derived from mass spectrometry data are Figs. 2a–k, 3a, 4a, 4c, 5c,d, 6b–j, and Extended Data Figs. 2d, 3a, 3c-e, 4a-e, 5b-f, 6a-e, 7b,c, 6e,f, 10b,c,f. All other supporting data are available as source data with this article. Source data are provided with this paper.
Code availability
Codes to reproduce analyses used in this study have been deposited at https://github.com/khavarilab/prober-manuscript and https://doi.org/10.5281/zenodo.6600968.
References
Cozzolino, F., Iacobucci, I., Monaco, V. & Monti, M. Protein-DNA/RNA interactions: an overview of investigation methods in the -omics era. J. Proteome Res. 20, 3018–3030 (2021).
Jutras, B. L., Verma, A. & Stevenson, B. Identification of novel DNA-binding proteins using DNA-affinity chromatography/pull down. Curr. Protoc. Microbiol. Chapter 1, Unit1F.1 (2012).
Liu, X. et al. In situ capture of chromatin interactions by biotinylated dCas9. Cell 170, 1028–1043 (2017).
Byrum, S. D., Taverna, S. D. & Tackett, A. J. Purification of a specific native genomic locus for proteomic analysis. Nucleic Acids Res. 41, e195 (2013).
Guillen-Ahlers, H. et al. HyCCAPP as a tool to characterize promoter DNA–protein interactions in Saccharomyces cerevisiae. Genomics 107, 267–273 (2016).
Fujita, T. & Fujii, H. Identification of proteins associated with an IFNγ-responsive promoter by a retroviral expression system for enChIP using CRISPR. PLoS One 9, e103084 (2014).
Déjardin, J. & Kingston, R. E. Purification of proteins associated with specific genomic loci. Cell 136, 175–186 (2009).
Mohammed, H. et al. Rapid immunoprecipitation mass spectrometry of endogenous proteins (RIME) for analysis of chromatin complexes. Nat. Protoc. 11, 316–326 (2016).
Rafiee, M. R. & Krijgsveld, J. Using ChIP-SICAP to identify proteins that co-localize in chromatin. Methods Mol. Biol. 2351, 275–288 (2021).
Schmidtmann, E., Anton, T., Rombaut, P., Herzog, F. & Leonhardt, H. Determination of local chromatin composition by CasID. Nucleus 7, 476–484 (2016).
Qiu, W. et al. Determination of local chromatin interactions using a combined CRISPR and peroxidase APEX2 system. Nucleic Acids Res. 47, e52 (2019).
Myers, S. A. et al. Discovery of proteins associated with a predefined genomic locus via dCas9-APEX-mediated proximity labeling. Nat. Methods 15, 437–439 (2018).
Gao, X. D. et al. C-BERST: defining subnuclear proteomic landscapes at genomic elements with dCas9-APEX2. Nat. Methods 15, 433–436 (2018).
Ummethum, H. & Hamperl, S. Proximity labeling techniques to study chromatin. Front. Genet. 11, 450 (2020).
Ramanathan, M. et al. RNA–protein interaction detection in living cells. Nat. Methods 15, 207–212 (2018).
Caygill, E. E. & Brand, A. H. The GAL4 system: a versatile system for the manipulation and analysis of gene expression. Methods Mol. Biol. 1478, 33–52 (2016).
Wobbe, C. R. et al. In vitro replication of DNA containing either the SV40 or the polyoma origin. Philos. Trans. R. Soc. Lond. B Biol. Sci. 317, 439–453 (1987).
Teo, G. et al. SAINTq: scoring protein–protein interactions in affinity purification–mass spectrometry experiments with fragment or peptide intensity data. Proteomics 16, 2238–2245 (2016).
Mellacheruvu, D. et al. The CRAPome: a contaminant repository for affinity purification–mass spectrometry data. Nat. Methods 10, 730–736 (2013).
Choi, H. et al. Analyzing protein–protein interactions from affinity purification–mass spectrometry data with SAINT. Curr. Protoc. Bioinformatics Chapter 8, Unit8.15 (2012).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Cai, Y. et al. YY1 functions with INO80 to activate transcription. Nat. Struct. Mol. Biol. 14, 872–874 (2007).
Davis, C. A. et al. The Encyclopedia of DNA Elements (ENCODE): data portal update. Nucleic Acids Res. 46, D794–D801 (2018).
Sloan, C. A. et al. ENCODE data at the ENCODE portal. Nucleic Acids Res. 44, D726–D732 (2016).
Singh, B. & Nath, S. K. Identification of proteins interacting with single nucleotide polymorphisms (SNPs) by DNA pull-down assay. Methods Mol. Biol. 1855, 355–362 (2019).
Chen, E. Y. et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128 (2013).
Roux, K. J., Kim, D. I., Raida, M. & Burke, B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 196, 801–810 (2012).
Tehranchi, A. K. et al. Pooled ChIP-seq links variation in transcription factor binding to complex disease risk. Cell 165, 730–741 (2016).
Nica, A. C. & Dermitzakis, E. T. Expression quantitative trait loci: present and future. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368, 20120362 (2013).
Stacey, S. N. et al. New basal cell carcinoma susceptibility loci. Nat. Commun. 6, 6825 (2015).
Yan, J. et al. Systematic analysis of binding of transcription factors to noncoding variants. Nature 591, 147–151 (2021).
Chiba, K. et al. Cancer-associated TERT promoter mutations abrogate telomerase silencing. Elife 4, e07918 (2015).
Bell, R. J. et al. Cancer. The transcription factor GABP selectively binds and activates the mutant TERT promoter in cancer. Science 348, 1036–1039 (2015).
Makowski, M. M. et al. An interaction proteomics survey of transcription factor binding at recurrent TERT promoter mutations. Proteomics 16, 417–426 (2016).
Heidenreich, B. & Kumar, R. TERT promoter mutations in telomere biology. Mutat. Res. Rev. Mutat. Res. 771, 15–31 (2017).
Weintraub, A. S. et al. YY1 is a structural regulator of enhancer–promoter loops. Cell 171, 1573–1588 (2017).
Zhang, W. et al. A global transcriptional network connecting noncoding mutations to changes in tumor gene expression. Nat. Genet. 50, 613–620 (2018).
Uffelmann, E. et al. Genome-wide association studies. Nat. Rev. Methods Primers 1, 59 (2021).
Tewhey, R. et al. Direct identification of hundreds of expression-modulating variants using a multiplexed reporter assay. Cell 165, 1519–1529 (2016).
Ulirsch, J. C. et al. Systematic functional dissection of common genetic variation affecting red blood cell traits. Cell 165, 1530–1545 (2016).
Acknowledgements
This work was supported by the US Veterans Affairs Office of Research and Development I01BX00140908, National Institutes of Health, National Institute for Arthritis and Musculoskeletal and Skin Diseases (NIH/NIAMS) AR076965 and AR049737, and by the NIH National Human Genome Research Institute (NIH/NHGRI) HG010856 (P.A.K.). The authors thank R. Leib at Stanford University Mass Spectrometry for help with mass spectrometry, with core support from NIH P30 CA124435 and S10RR027425.
Author information
Authors and Affiliations
Contributions
S.M. designed and performed experiments, analyzed data and wrote the manuscript. W.M., D.R., V.L.-P., Z.S., M.R., D.L.R., P.N., M.G., L.M.M., Y.-Y.Y., L.L. and Y.W. contributed to designing and performing the experiments. R.M.M. analyzed the data. Y.Z., D.F.P. and I.F. provided additional computational support. P.A.K. supervised the project, designed experiments, analyzed data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Methods thanks Jian Yan, Markku Varjosalo and Samuel Myers for their contribution to the peer review of this work. Primary Handling Editor: Lei Tang, in collaboration with the Nature Methods team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 PROBER specificity and optimization of motif concatemers, insert size and bait copy number.
a, PROBER-WB of canonical YY1, NF-κB, STAT1 DNA motifs and their nucleotide composition-matched scrambled controls + /- TNF-α or IFN-γ probed together with YY1, RelA, or STAT1 antibodies. b, Heatmap of a representing fold enrichment of signals, n = 1 biological replicate. c, PROBER-WB showing effect of single-copy, duplicate, triplicate, quadruplicate and quintuplicate YY1 motifs on fold enrichment, d, quantification of c representing mean enrichment, n = 2 biologically independent replicates. e, Trimeric NF-κB motif and matched scrambled sequences were cloned in pBait with different lengths of flanks at both ends to obtain inserts of varying sizes. The NF-κB motifs are shown as “N”, position of UAS elements are also shown. f, PROBER-WB showing the effect of insert size on RelA enrichment, and g, quantification of f representing mean enrichment, n = 2 biologically independent replicates. Note that increasing distance between NF-κB motifs and UAS beyond 25-nt stuffer flanks results in loss of enrichment because it puts the TFs outside BASU labeling radius. h, Nuclear plasmid copies after transfection of HEK293T cells with varying amounts of bait plasmid and 3 μg pSprayer; an SV40 Ori-less derivative of pBait with trimeric YY1 motif was used eliminate copy number increase by endogenous large T antigen, and i, resulting YY1 PROBER fold enrichment in response to bait copy number in h. n = 1 biological replicate.
Extended Data Fig. 2 Validation of YY1 interactors.
a, PROBER-WB of YY1 motif showing co-enrichment of ZHX1, ZHX2 and ZHX3, and b, quantification of enrichment normalized to BASU-Gal4DBD, n = 1 biological replicate. c, WB of normal and anti-YY1 siRNA treated HEK293T cells. Bar plot indicates YY1 levels normalized to β-actin, n = 2 biologically independent replicates. d, logFC (between control vs. YY1-knockdown condition) vs. average FC (over scramble controls) plot representing effect of YY1 knockdown on YY1 PROBER. Color codes are as indicated in Fig. 2a. e, Euler diagram and UpSet plot of overlapping ENCODE ChIP-seq peaks in HepG2 cells. f, Euler diagram and UpSet plot of overlapping ENCODE ChIP-seq peaks in K562 cells. In both UpSet plots, bottom panels indicate the combination of peak sets, and the number of peaks in each set. The middle panel indicates the size of the overlapping sets. The top panel indicates the relative enrichment of each combination.
Extended Data Fig. 3 Detection and validation of RelA and STAT1 interactors.
a, logFC (between + TNF-α vs. -TNF-α condition) vs. average FC (over scramble controls) plot representing effect of TNF-α stimulation on NF-κB PROBER. Color codes are as indicated in Fig. 2a. b, WB of normal and anti-RelA siRNA treated HEK293T cells in presence of TNF-α. Bar plot indicates RelA levels normalized to β-actin, n = 2 biologically independent replicates. c, logFC (between control vs. RelA-knockdown condition) vs. average FC (over scramble controls) plot representing effect of RelA knockdown on NF-κB PROBER. Color codes are as indicated before. d, Differential analysis of proteins enriched in STAT1 PROBER-MS in –IFN-γ vs. +IFN-γ conditions. Red and cyan dots are as indicated before, pink dots indicate select DNA/chromatin binders with FDR above 0.25. e, logFC (between + IFN-γ vs. -IFN-γ condition) vs. average FC (over scramble controls) plot representing effect of IFN-γ stimulation on STAT1 PROBER, color codes as indicated in d. f, PWM of ETS and STAT1 motifs (Source: http://jaspar.genereg.net). Sequence of STAT1 motif used in PROBER is shown at the bottom.
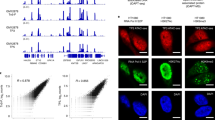
Extended Data Fig. 4 PROBER-MS reflects endogenous DNA–protein interactions.
a-c, SAINT score vs. fold-change (FC) scatter plots of endogenous BS1, BS2 and BS3 PROBER-MS, n = 3 biologically independent replicates. The 26-nt chromosomal sequences that were cloned in triplicate are shown on top of each plot, the central YY1 motifs are indicated in red. The ETS motif overlapping YY1 motif in BS3 is underlined. Proteins that were enriched (SAINT scores ≥ 0.9) are indicated in red (known DNA or chromatin binder) or cyan (not known to be a DNA or chromatin binder); known YY1 interactors are highlighted in red. d-f, NFRKB, GABPA and ELK1 ChIP-seq signal p-values at BS1, BS2 and BS3 from publicly available datasets, Integrated Genome Viewer (IGV) (https://software.broadinstitute.org/software/igv/) screenshots of approximately 1 kb region surrounding BS1, BS2 and BS3 are shown.
Extended Data Fig. 5 PROBER-WB in multiple cell lines and primary cells.
a, PROBER-WB of YY1 motif in U2OS (Bone osteosarcoma), A549 (lung carcinoma), ReNcell® CX (immortalized human neural progenitor), primary keratinocytes (NHEK), and primary dermal fibroblasts. Bar plots represent mean YY1-fold enrichment, n = 3 (for U2OS and ReNCell CX) and 2 (for A549, NHEK and fibroblasts) biologically independent replicates. b, SAINT score vs. fold-change (FC) scatter plots of triplicate YY1 motif (used in Fig. 1b), and c, triplicate 26-nt endogenous BS1 loci (used in Extended Data Fig. 4a) from U2OS cells, n = 3 biologically independent PROBER-MS replicates. Color codes are as described in Fig. 2a. d, Venn diagrams showing proteins enriched at triplicate YY1 motif and endogenous BS1 construct in U2OS cells and e, endogenous BS1 construct in HEK293T and U2OS cells. f, Differential analysis of proteins enriched at BS1 from HEK293T and U2OS PROBER-MS, color codes as indicated before. Dotted line represents FDR 0.25. g, Comparison of biotinylation (green smear) in HEK293T and U2OS cells in cell lysate and after enrichment by streptavidin pull-down. Note that BASU-GAL4 is minimally expressed in both cell types and visible in WB only after pull-down.
Extended Data Fig. 6 DNA affinity pull-down using an NF-κB motif.
a, SAINT score vs. fold-change (FC) scatter plots of DNA affinity pull-down with triplicate NF-κB motif from nuclear lysates of TNF-α stimulated and b, unstimulated HEK293T cells, n = 2 biologically independent replicates, color codes are as indicated in Fig. 2a. NF-κB family proteins are indicated in red text. c, Scatter plot comparing fold-change (FC) of proteins enriched (SAINT ≥ 0.9) by pull-down in TNF-α stimulated vs. unstimulated conditions. Blue dots indicate proteins enriched with TNF-α, green dots indicate proteins enriched in unstimulated condition, and red dots indicate proteins enriched under both conditions. d, Differential analysis of proteins enriched in NF-κB DNA affinity pull-down +TNF-α vs. -TNF-α, color codes are as indicated in a. Horizontal dotted line indicates FDR 0.25. e, PROBER vs. DNA pull-down (–TNF-α) fold-change scatter plot; blue dots indicate proteins enriched (SAINT ≥ 0.9) by PROBER, green dots indicate proteins enriched by DNA pull-down, and red dots indicate proteins enriched by both methods.
Extended Data Fig. 7 RelA BioID using BASU-RelA fusion.
a, Expression of BASU-RelA fusion protein compared to endogenous RelA, n = 1 biological replicate. b, SAINT score vs. FC scatter plots of RelA BioID in presence and absence of TNF-α, n = 3 biologically independent replicates. Color codes are as indicated in Fig. 2a, NF-κB family proteins are indicated in red text. c, PROBER vs. BioID (–TNF-α) fold-change scatter plot; blue dots indicate proteins enriched (SAINT ≥ 0.9) by PROBER, green dots indicate proteins enriched by DNA pull-down, and red dots indicate proteins enriched by both methods. d, Proximity Ligation Assay (PLA) of RelA and PHF21A in presence or absence of TNF-α. Nuclei are visualized with DAPI (blue), orange dots indicate PLA signal. The boxplot represents nuclear PLA signal obtained by counting orange dots in 8 representative transfected cells, the center is the median, lower and upper limits depict the first and third quartile, and the whiskers show minimum and maximum dot counts. ***P < 0.001, 4.03 × 10−4 (obtained by two-tailed Student’s t-test from 8 transfected cells, n = 1 biological replicate). e, Scatter plot comparing fold-change (FC) of proteins enriched (SAINT ≥ 0.9) by BioID in TNF-α stimulated vs. unstimulated conditions. Blue dots indicate proteins enriched with TNF-α, green dots indicate proteins enriched in unstimulated condition, and red dots indicate proteins enriched under both conditions. f, Differential analysis of proteins enriched in RelA BioID +TNF-α vs. -TNF-α, color codes as indicated in b. Horizontal dotted line indicates FDR 0.25. NF-κB family proteins and common RelA interactors that were not differentially enriched are indicated in red text.
Extended Data Fig. 8 C-BERST WB of chromosomal YY1 binding sites.
a, C-BERST WB of centromeric α-satellite repeats as positive control showing enrichment of CENP-B and corresponding streptavidin blot, n = 2 biologically independent replicates. b, anti-FLAG ChIP-qPCR of BS1, BS2, BS3 loci and a gene desert negative control locus showing specific recruitment of dCas9-APEX2-mCherry fusion protein using sgBS1, sgBS2, and sgBS3. Tukey’s multiple comparison test performed, ns= not significant, ***P < 0.001, n = 2 biologically independent replicates. c, C-BERST WB of BS1, BS2, and BS3 showing no YY1 enrichment over sgNS control. Bar plots represent mean enrichment of 2 independent replicates.
Extended Data Fig. 9 Detection of differential TF recruitment at SNPs.
a, Sequence surrounding SNP rs2349075 showing both A- and G-alleles, and expression of CASP8 associated with rs2349075 G allele in sun exposed lower leg skin (Source- http://www.gtexportal.org/home/). The c-Jun motif overlapping rs2349075 is underlined. b, Luciferase assay of SNP rs2349075 A-and G-alleles showing higher reporter activity associated with G allele with phorbol ester treatment. Bar plots represent mean normalized expressions, n = 2 biologically independent replicates. Two-tailed Student’s t-test, *P < 0.05, 1.91 × 10−2. c, PROBER-WB of SNPs rs2349075 A-and G- alleles in presence of PMA. Bar plots represent mean enrichment, n = 3 biologically independent replicates. Two-tailed Student’s t-test performed; *P < 0.05, 1.54 × 10−2. d, Sequence surrounding SNP rs7132503 showing both A- and G-alleles, and expression of linked pseudogene RP1-102E24.10 in pancreas tissue (http://www.gtexportal.org/home/). The overlapping NF-κB motif is underlined. e, PROBER-WB of SNPs rs7132503 both A-and G- alleles in presence of TNF-α, bar plot represents mean enrichment, n = 2 biologically independent replicates. Two-tailed Student’s t-test performed, *P < 0.05, 4.86 × 10−2. f, Episomal ChIP-qPCRs performed on plasmids encoding approximately 1 kb native chromosomal region surrounding G-or A-alleles of rs7132503 in presence of TNF-α. Bar plots represent mean enrichment, n = 4 biologically independent replicates. Two-tailed Student’s t-test performed, ***P < 0.001, 3.47 × 10−6. g, PROBER-WB on c-Jun binding SNP variants identified by SNP-SELEX. Normalized PROBER enrichment scores and area under the curve (AUC) values derived by SNP-SELEX experiment are indicated below.
Extended Data Fig. 10 PROBER with WT hTERT promoter and high-throughput compatibility.
a, Presence of ZNF148 and partial ZNF282 motifs in hTERT promoter, PWMs were downloaded from http://jaspar.genereg.net. b, PROBER-MS with chr5:1,295,260-1,295,218 region (single-copy native fragment) of hTERT promoter, color coding as indicated in Fig. 2a. c, PROBER-MS predicted interactors of chr5:1,295,260-1,295,218 region, DNA/chromatin binders are indicated in blue. d, siRNA-mediated knockdown of HDAC2, RBBP4, ZNF148 and ZNF281 in HEK293T cells. Bar plots represent mean relative expression, n = 3 biologically independent replicates, error bars represent SEM. e, TERT expression from WT hTERT loci in HEK293T after siRNA-mediated knockdown of HDAC2, RBBP4, ZNF148 and ZNF281. Bar plots represent mean relative expression, n = 3 biologically independent replicates, error bars represent SEM. Two-tailed t-test performed with Welch’s correction; *P < 0.05, 2.13 × 10−2 (RBBP4) and 1.10 × 10−2 (HDAC2); **P < 0.01, 8.76 × 10−3 (ZNF148) and 3.59 × 10−3 (ZNF281). f, SAINT score vs. fold-change (FC) scatter plot resulting from PROBER-MS of triplicate YY1 motif (used in Fig. 1b) in HEK293T cells, where MS samples were prepared by high-throughput compatible on-bead sample processing (gel-free purification). Color codes are as described in b. g, Venn-diagram showing proteins enriched at triplicate YY1 motif in PROBER-MS samples prepared by gel-based or on-bead processing.
Supplementary information
Source data
Source Data Fig. 1
Unprocessed Western Blots and Statistical Source Data
Source Data Fig. 1
Unprocessed Western Blots and Statistical Source Data
Source Data Fig. 2
Statistical Source Data
Source Data Fig. 3
Statistical Source Data
Source Data Fig. 4
Statistical Source Data
Source Data Fig. 5
Unprocessed Western Blots and Statistical Source Data
Source Data Fig. 5
Unprocessed Western Blots and Statistical Source Data
Source Data Fig. 6
Statistical Source Data
Source Data Extended Data Fig. 1
Unprocessed Western Blots and Statistical Source Data
Source Data Extended Data Fig. 1
Unprocessed Western Blots and Statistical Source Data
Source Data Extended Data Fig. 2
Unprocessed Western Blots and Statistical Source Data
Source Data Extended Data Fig. 2
Unprocessed Western Blots and Statistical Source Data
Source Data Extended Data Fig. 3
Unprocessed Western Blots and Statistical Source Data
Source Data Extended Data Fig. 3
Unprocessed Western Blots and Statistical Source Data
Source Data Extended Data Fig. 4
Statistical Source Data
Source Data Extended Data Fig. 5
Unprocessed Western Blots and Statistical Source Data
Source Data Extended Data Fig. 5
Unprocessed Western Blots and Statistical Source Data
Source Data Extended Data Fig. 6
Statistical Source Data
Source Data Extended Data Fig. 7
Unprocessed Western Blots and Statistical Source Data
Source Data Extended Data Fig. 7
Unprocessed Western Blots and Statistical Source Data
Source Data Extended Data Fig. 8
Unprocessed Western Blots and Statistical Source Data
Source Data Extended Data Fig. 8
Unprocessed Western Blots and Statistical Source Data
Source Data Extended Data Fig. 9
Unprocessed Western Blots and Statistical Source Data
Source Data Extended Data Fig. 9
Unprocessed Western Blots and Statistical Source Data
Source Data Extended Data Fig. 10
Statistical Source Data
Rights and permissions
About this article
Cite this article
Mondal, S., Ramanathan, M., Miao, W. et al. PROBER identifies proteins associated with programmable sequence-specific DNA in living cells. Nat Methods 19, 959–968 (2022). https://doi.org/10.1038/s41592-022-01552-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-022-01552-w