Abstract
Transcriptomic data is often affected by uncontrolled variation among samples that can obscure and confound the effects of interest. This variation is frequently due to unintended differences in developmental stages between samples. The transcriptome itself can be used to estimate developmental progression, but existing methods require many samples and do not estimate a specimen’s real age. Here we present real-age prediction from transcriptome staging on reference (RAPToR), a computational method that precisely estimates the real age of a sample from its transcriptome, exploiting existing time-series data as reference. RAPToR works with whole animal, dissected tissue and single-cell data for the most common animal models, humans and even for non-model organisms lacking reference data. We show that RAPToR can be used to remove age as a confounding factor and allow recovery of a signal of interest in differential expression analysis. RAPToR will be especially useful in large-scale single-organism profiling because it eliminates the need for accurate staging or synchronisation before profiling.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Source data for all figures is provided. Source data are provided with this paper.
Code availability
The code to download and (pre)process the data, perform the analyses and generate the figures of this paper can be found at https://gitbio.ens-lyon.fr/LBMC/qrg/raptor-analysis
References
Francesconi, M. & Lehner, B. Reconstructing and analysing cellular states, space and time from gene expression profiles of many cells and single cells. Mol. Biosyst. 11, 2690–2698 (2015).
Risso, D., Ngai, J., Speed, T. P. & Dudoit, S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat. Biotechnol. 32, 896–902 (2014).
Leek, J. T. & Storey, J. D. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 3, e161 (2007).
Stegle, O., Parts, L., Piipari, M., Winn, J. & Durbin, R. Using probabilistic estimation of expression residuals (PEER) to obtain increased power and interpretability of gene expression analyses. Nat. Protoc. 7, 500–507 (2012).
Gómez-Orte, E. et al. Effect of the diet type and temperature on the C. elegans transcriptome. Oncotarget 9, 9556–9571 (2018).
MacNeil, L. T., Watson, E., Arda, H. E., Zhu, L. J. & Walhout, A. J. Diet-induced developmental acceleration independent of TOR and insulin in C. elegans. Cell 153, 240–252 (2013).
Ludewig, A. H. et al. Larval crowding accelerates C. elegans development and reduces lifespan. PLoS Genet. 13, e1006717 (2017).
Kuntz, S. G. & Eisen, M. B. Drosophila embryogenesis scales uniformly across temperature in developmentally diverse species. PLoS Genet. 10, e1004293 (2014).
Perez, M. F., Francesconi, M., Hidalgo-Carcedo, C. & Lehner, B. Maternal age generates phenotypic variation in Caenorhabditis elegans. Nature 552, 106–109 (2017).
Snoek, L. B. et al. A rapid and massive gene expression shift marking adolescent transition in C. elegans. Sci Rep. 4, 3912 (2014).
Rockman, M. V., Skrovanek, S. S. & Kruglyak, L. Selection at linked sites shapes heritable phenotypic variation in C. elegans. Science 330, 372–376 (2010).
Francesconi, M. & Lehner, B. The effects of genetic variation on gene expression dynamics during development. Nature 505, 208–211 (2014).
Hibbs, M. A. et al. Exploring the functional landscape of gene expression: directed search of large microarray compendia. Bioinformatics 23, 2692–2699 (2007).
Lu, P., Nakorchevskiy, A. & Marcotte, E. M. Expression deconvolution: a reinterpretation of DNA microarray data reveals dynamic changes in cell populations. Proc. Natl Acad. Sci. 100, 10370–10375 (2003).
O’Duibhir, E. et al. Cell cycle population effects in perturbation studies. Mol. Syst. Biol. 10, 732 (2014).
Street, K. et al. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics 19, 477 (2018).
Haghverdi, L., Büttner, M., Wolf, F. A., Buettner, F. & Theis, F. J. Diffusion pseudotime robustly reconstructs lineage branching. Nat. Methods 13, 845–848 (2016).
Trapnell, C. et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014).
Anavy, L. et al. BLIND ordering of large-scale transcriptomic developmental timecourses. Development 141, 1161–1166 (2014).
Kim, Dhyun, Grün, D. & van Oudenaarden, A. Dampening of expression oscillations by synchronous regulation of a microRNA and its target. Nat. Genet. 45, 1337–1344 (2013).
Meeuse, M. W. et al. Developmental function and state transitions of a gene expression oscillator in Caenorhabditis elegans. Mol. Syst. Biol. 16, e9498 (2020).
Reinke, V., San Gil, I., Ward, S. & Kazmer, K. Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development 131, 311–323 (2004).
Domazet-Lošo, T. & Tautz, D. A phylogenetically based transcriptome age index mirrors ontogenetic divergence patterns. Nature 468, 815–818 (2010).
Xue, L. et al. Global expression profiling reveals genetic programs underlying the developmental divergence between mouse and human embryogenesis. BMC Genomics 14, 568 (2013).
Graveley, B. R. et al. The developmental transcriptome of Drosophila melanogaster. Nature 471, 473–479 (2011).
Hendriks, G.-J., Gaidatzis, D., Aeschimann, F. & Großhans, H. Extensive oscillatory gene expression during C. elegans larval development. Mol. Cell 53, 380–392 (2014).
Levin, M. et al. The mid-developmental transition and the evolution of animal body plans. Nature 531, 637–641 (2016).
Rauwerda, H. et al. Transcriptome dynamics in early zebrafish embryogenesis determined by high-resolution time course analysis of 180 successive, individual zebrafish embryos. BMC Genomics 18, 287 (2017).
Collins, J. E. et al. Common and distinct transcriptional signatures of mammalian embryonic lethality. Nat. Commun. 10, 2792 (2019).
Somel, M., Khaitovich, P., Bahn, S., Pääbo, S. & Lachmann, M. Gene expression becomes heterogeneous with age. Curr. Biol. 16, R359–R360 (2006).
Kedlian, V. R., Donertas, H. M. & Thornton, J. M. The widespread increase in inter-individual variability of gene expression in the human brain with age. Aging 11, 2253–2280 (2019).
Martinez-Jimenez, C. P. et al. Aging increases cell-to-cell transcriptional variability upon immune stimulation. Science 355, 1433–1436 (2017).
Hou, L. et al. A systems approach to reverse engineer lifespan extension by dietary restriction. Cell Metab. 23, 529–540 (2016).
Golden, T. R., Hubbard, A., Dando, C., Herren, M. A. & Melov, S. Age-related behaviors have distinct transcriptional profiles in Caenorhabditis elegans. Aging Cell 7, 850–865 (2008).
Pletcher, S. D. et al. Genome-wide transcript profiles in aging and calorically restricted Drosophila melanogaster. Curr. Biol. 12, 712–723 (2002).
Chen, C.-Y. et al. Effects of aging on circadian patterns of gene expression in the human prefrontal cortex. Proc. Natl Acad. Sci. 113, 206–211 (2016).
Pantalacci, S. et al. Transcriptomic signatures shaped by cell proportions shed light on comparative developmental biology. Genome Biol. 18, 29 (2017).
Sémon, M. et al. Comparison of developmental genome expression in rodent molars reveals extensive developmental system drift. Preprint at bioRxiv https://doi.org/10.1101/2020.04.22.043422 (2020).
Petropoulos, S. et al. Single-cell RNA-seq reveals lineage and X chromosome dynamics in human preimplantation embryos. Cell 165, 1012–1026 (2016).
Perez, M. F. Neuronal perception of the social environment generates an inherited memory that controls the development and generation time of C. elegans. Curr. Biol. 31, 4256–4268 (2021).
Kalinka, A. T. et al. Gene expression divergence recapitulates the developmental hourglass model. Nature 468, 811–814 (2010).
Deng, Q., Ramsköld, D., Reinius, B. & Sandberg, R. Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science 343, 193–196 (2014).
Vassena, R. et al. Waves of early transcriptional activation and pluripotency program initiation during human preimplantation development. Development 138, 3699–3709 (2011).
Cuthbert, J. M. et al. Comparing mRNA and sncRNA profiles during the maternal-to-embryonic transition in bovine IVF and scNT embryos. Biol. Reprod. 105, 1401–1415 (2021).
Li, J. J., Huang, H., Bickel, P. J. & Brenner, S. E. Comparison of D. melanogaster and C. elegans developmental stages, tissues, and cells by modENCODE RNA-seq data. Genome Res. 24, 1086–1101 (2014).
Lewis, J. A., Szilagyi, M., Gehman, E., Dennis, W. E. & Jackson, D. A. Distinct patterns of gene and protein expression elicited by organophosphorus pesticides in Caenorhabditis elegans. BMC Genomics 10, 202 (2009).
Lehrbach, N. J. et al. Post-developmental microRNA expression is required for normal physiology, and regulates aging in parallel to insulin/IGF-1 signaling in C. elegans. RNA 18, 2220–2235 (2012).
Hall, S. E., Beverly, M., Russ, C., Nusbaum, C. & Sengupta, P. A cellular memory of developmental history generates phenotypic diversity in C. elegans. Curr. Biol. 20, 149–155 (2010).
Miki, T. S., Carl, S. H. & Großhans, H. Two distinct transcription termination modes dictated by promoters. Genes Dev. 31, 1870–1879 (2017).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47–e47 (2015).
Borchani, H., Varando, G., Bielza, C. & Larrañaga, P. A survey on multi-output regression. WIRES Data Min. Knowl. 5, 216–233 (2015).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A. & Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017).
Hill, M. A. Mouse Stages. Embryology https://embryology.med.unsw.edu.au/embryology/index.php/Main_Page (2022).
Michaelson, J. J., Alberts, R., Schughart, K. & Beyer, A. Data-driven assessment of eQTL mapping methods. BMC Genomics 11, 502 (2010).
Raudvere, U. et al. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 47, W191–W198 (2019).
Llobat, L. Pluripotency and growth factors in early embryonic development of mammals: a comparative approach. Vet. Sci. 8, 78 (2021).
Acknowledgements
We are grateful to S. E. Hall, M. Sémon, and S. Pantalacci for providing data from their profiling experiments. We are also grateful to G. Yvert, D. Jost, M. Sémon, A. Piazza, S. Pantalacci, and B. Lehner for their critical reading of the manuscript. M.F. is supported by INSERM. Work in the laboratory of M.F. is supported by a grant from the Agence Nationale pour la Recherche (ANR-19-CE12-0009 ‘InterPhero’), Université de Lyon (IDEX IMPULSION G19002CC) and ENS-Lyon (Projet emergent 2019). R.B. PhD fellowship is funded by the French Ministry of Research.
Author information
Authors and Affiliations
Contributions
M.F. and R.B. conceived the method; R.B. developed the computational framework and performed the analyses; and M.F. and R.B. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors report no competing interests.
Peer review
Peer review information
Nature Methods thanks Helge Grosshans, Adam Alexander Thil Smith and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Rita Strack, in collaboration with the Nature Methods team. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 RAPToR estimates fit gene expression data better than chronological age.
a, RAPToR estimates of D. melanogaster single-embryo samples27 staged on a reference built from bulk data25 plotted against established BLIND ranks27. b, Percentage of genes better fitted by either RAPToR estimates or chronological age modeled with splines using 2-8 degrees of freedom in otherwise identical models. c, R² of models from (b) gene count in each half of the plot is indicated in the corners. d,e, Principal components plotted along chronological age (d), and RAPToR estimates (e) (as in Fig. 2d-f).
Extended Data Fig. 2 Reference interpolation allows RAPToR estimates at high resolution.
a, RAPToR estimates of a zebrafish embryonic time-series from 9 spawns28 staged on a reference built from Domazet et al. data23 plotted against original developmental ranks28. b, First 2 principal components of the zebrafish time-series plotted against RAPToR age estimates. Spawns are color-coded. c,d, RAPToR estimates of the zebrafish time-series on the non-interpolated reference (i.e the sampling time of the reference sample with the highest correlation) vs. original developmental ranks (c) and vs. standard RAPToR estimates (as in a) (d). In a,c,d, original reference time points within the plot area are shown on the right, in blue.
Extended Data Fig. 3 Tissue-specific staging yields soma and germline ages.
a, RAPToR estimates of C. elegans Recombinant Inbred Lines (RILs)11 staged on the larval to young-adult reference built from Meeuse et al.21 vs. Francesconi & Lehner12 estimates. b-d, Comparison of RAPToR estimates of global age vs. germline age (b), global age vs. soma age (c), and soma age vs. germline age (d). e, Distribution of soma–germline heterochrony.
Extended Data Fig. 4 A delayed germline and an advanced soma.
a, Independent Components from ICA on C. elegans Recombinant Inbred Lines (RILs)11 joined to the (non-interpolated) reference data21 plotted along chronological age and RAPToR global estimates for the reference (orange) and RILs (black) respectively. b, Gene loadings on ICA components for all genes (n = 14132), germline genes (oogen. n = 582, sperm. n = 596) and soma (n = 2005) categories. Each box within violins spans the interquartile range (IQR), the central white dot denotes the median, and whiskers extend to 1.5×IQR in either direction. Category enrichment p-values derive from a two-sided hypergeometric test on genes with absolute loadings above 1.96. From left to right, p-values are IC2: p > 0.99, p < 1e-10, and p > 0.99; IC3: p < 1e-10, p = 2.66e-06, and p = 0.022; IC4: p > 0.99, p > 0.99, and p < 1e-10; IC5: p > 0.99, p > 0.99, and p < 1e-10; IC6: p < 1e-10, p > 0.99, and p = 6.54e-04; IC7: p > 0.99, p > 0.99, and p < 1e-10; IC8: p > 0.99, p > 0.99, and p < 1e-10. c,d, Summed (c) and per-component (d) Root Mean Square Error (RMSE) between RILs and reference fit on IC2-IC8 when shifting RIL (global) age estimates. RMSE per-component shows heterochrony, with soma dynamics of RILs matching younger reference time and the reverse for germline dynamics. *: p < 0.05, **: p < 0.01, ***: p < 0.001.
Extended Data Fig. 5 Soma–germline heterochrony among C. elegans recombinant lines.
Recombinant Inbred Lines (RILs)11 are staged on the larval to young-adult reference built from Meeuse et al. samples21. a, Percentage of genes better fitted by either RAPToR global, soma, or germline age estimates, modeled with splines with 4, 6, or 8 degrees of freedom in otherwise identical models. Genes are classified into spermatogenesis, oogenesis, somatic, or other (see methods). b, R² per gene of models with global, soma, or germline age estimates as predictors for 4, 6, and 8 spline degrees of freedom.
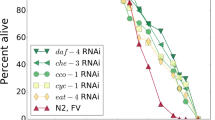
Extended Data Fig. 6 RAPToR age estimates synchronize expression dynamics across species.
Extended Data Fig. 7 Staging M. musculus single cells on H. sapiens reference.
Single cells from M. musculus embryos42 were staged on a H. sapiens single-cell embryogenesis reference39 using orthologs. a, First 2 principal components of a PCA done on the 1000 most variable genes. A principal curve is fit on the first 3 components. Cells are colored by RAPToR age estimate on the H. sapiens reference. b, RAPToR age estimates of M. musculus single cells on H. sapiens reference vs. cell ranks along principal curve (a). c, Chronological age of M. musculus single cells vs. RAPToR age estimates on H. sapiens reference using top 10% most correlated genes between mouse and human for staging (see methods). d, H. sapiens (red) and M. musculus (black) clustered gene expression profiles (aggregated per time point) of highest-correlated genes between both species (see methods).
Extended Data Fig. 8 Staging C. elegans embryogenesis with D. melanogaster.
a, C. elegans embryo samples from Levin et al.27 staged on the D. melanogaster reference built from Graveley et al.25 samples. Gaps appear in the estimates, likely at points where fly expression dynamics are incompatible with those of worms. b, As in (a), staging on the adjusted fly reference and using top 10% most correlated genes between fly and worm embryogenesis (see methods). c, D. melanogaster (red) and C. elegans (black) clustered gene expression profiles of highest-correlated genes between both species (see methods). d, ICA components of the C. elegans embryo time course plotted along sampling time. Both the red highlighted outlier and 4 samples with erroneous chronological age (circled in IC1) are omitted from analysis (see methods).
Extended Data Fig. 9 Estimating the impact of development by integrating reference data.
a-c, Cartoon detailing how the log-fold-changes (logFCs) of a differential expression analysis between two sample groups (a) and the logFCs of their matching time points in the RAPToR interpolated reference (b) can be compared to quantify the impact of development (c).
Extended Data Fig. 10 Correcting the effect of development by integrating reference data.
Samples from C. elegans time-course experiments of wildt-type (WT) and xrn-2 mutants, profiled by Miki et al.49, and staged on the larval to young-adult reference built from Meeuse et al. samples21, are used to validate developmental correction approach (see also Fig. 5f-i). a, Cartoon of a model integrating a window of reference data, with Strain and Batch coefficients shown in blue. b, Number of DE genes found by a standard differential expression model (FDR < 0.05) increases with the age gaps between compared groups, with a quasi-constant fraction of truly DE genes. c, Area under PR curves (AUPRC) in detecting gold-standard DE genes for standard differential expression model p-value, age-corrected logFCs, or the age-corrected classifier for each shifted WT subset. d, w parameter optimization for shifted WT sets, by maximizing area under the PR curves. e, PR curves of gold-standard gene detection by the age-corrected classifier for each shifted WT subset. f, Correlation of expected development logFCs and observed logFCs between the xrn-2 subset and combinations of 3-sample WT sets (note these are not the “WT -n” subsets, see Supplementary Table 13). g, Relationship between optimal w and sample-reference logFC correlation, as in (f). h, Optimal spline degree-of-freedom (df) selection for the different WT shifted sets by reaching a residual Sum of Square (SSQ) plateau. The selected df increases with the shift, which is expected since the reference window to include gets larger and may thus contain more complex dynamics. DE, Differentially Expressed. logFC, log2 fold-change. FDR, false discovery rate, PR: Precision-Recall.
Supplementary information
Supplementary Information
Supplementary Notes 1, 2 and Supplementary Figures 1–14.
Supplementary Tables
Supplementary Tables 1–13.
Supplementary Data 1
All plot data for Supplementary Fig. 1 plots.
Supplementary Data 2
All plot data for Supplementary Fig. 2 plots.
Supplementary Data 3
All plot data for Supplementary Fig. 3 plots.
Supplementary Data 4
All plot data for Supplementary Fig. 4 plots.
Supplementary Data 5
All plot data for Supplementary Fig. 5 plots.
Supplementary Data 6
All plot data for Supplementary Fig. 6 plots.
Supplementary Data 7
All plot data for Supplementary Fig. 7 plots.
Supplementary Data 8
All plot data for Supplementary Fig. 8 plots.
Supplementary Data 9
All plot data for Supplementary Fig. 9 plots.
Supplementary Data 10
All plot data for Supplementary Fig. 10 plots.
Supplementary Data 11
All plot data for Supplementary Fig. 11 plots.
Supplementary Data 12
All plot data for Supplementary Fig. 12 plots.
Supplementary Data 13
All plot data for Supplementary Fig. 13 plots.
Supplementary Data 14
All plot data for Supplementary Fig. 14 plots.
Source data
Source Data Fig. 2
All plot data.
Source Data Fig. 3
All plot data.
Source Data Fig. 4
All plot data.
Source Data Fig. 5
All plot data (classifiers are given for ROC curves).
Source Data Extended Data Fig. 1
All plot data.
Source Data Extended Data Fig. 2
All plot data.
Source Data Extended Data Fig. 3
All plot data.
Source Data Extended Data Fig. 4
All plot data.
Source Data Extended Data Fig. 5
All plot data.
Source Data Extended Data Fig. 6
All plot data.
Source Data Extended Data Fig. 7
All plot data.
Source Data Extended Data Fig. 8
All plot data.
Source Data Extended Data Fig. 10
All plot data (classifiers are given for ROC curves).
Rights and permissions
About this article
Cite this article
Bulteau, R., Francesconi, M. Real age prediction from the transcriptome with RAPToR. Nat Methods 19, 969–975 (2022). https://doi.org/10.1038/s41592-022-01540-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-022-01540-0
This article is cited by
-
Mitochondrial aconitase suppresses immunity by modulating oxaloacetate and the mitochondrial unfolded protein response
Nature Communications (2023)
-
Biomarkers of aging
Science China Life Sciences (2023)
-
Transcriptional atlas of the human immune response to 13 vaccines reveals a common predictor of vaccine-induced antibody responses
Nature Immunology (2022)