Abstract
Regulation of receptor tyrosine kinase (RTK) activity is necessary for studying cell signaling pathways in health and disease. We developed a generalized approach for engineering RTKs optically controlled with far-red light. We targeted the bacterial phytochrome DrBphP to the cell surface and allowed its light-induced conformational changes to be transmitted across the plasma membrane via transmembrane helices to intracellular RTK domains. Systematic optimization of these constructs has resulted in optically regulated epidermal growth factor receptor, HER2, TrkA, TrkB, FGFR1, IR1, cKIT and cMet, named eDrRTKs. eDrRTKs induced downstream signaling in mammalian cells in tens of seconds. The ability to activate eDrRTKs with far-red light enabled spectral multiplexing with fluorescent probes operating in a shorter spectral range, allowing for all-optical assays. We validated eDrTrkB performance in mice and found that minimally invasive stimulation in the neocortex with penetrating via skull far-red light-induced neural activity, early immediate gene expression and affected sleep patterns.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The main data supporting the findings of this study are available within the article and its supplementary information. The main resultant plasmids encoding eDrRTKs are deposited in the Addgene depository (nos. 183994–184001). The code used in this study for LED switching is available at https://docs.arduino.cc/built-in-examples/basics/Blink. Source data are provided with this paper.
References
Lemmon, M. A. & Schlessinger, J. Cell signaling by receptor tyrosine kinases. Cell 141, 1117–1134 (2010).
Leopold, A. V. & Verkhusha, V. V. Light control of RTK activity: from technology development to translational research. Chem. Sci. 11, 10019–10034 (2020).
Grusch, M. et al. Spatio-temporally precise activation of engineered receptor tyrosine kinases by light. EMBO J. 33, 1713–1726 (2014).
Huang, P. et al. Optical activation of TrkB signaling. J. Mol. Biol. 432, 3761–3770 (2020).
Piatkevich, K. D., Subach, F. V. & Verkhusha, V. V. Engineering of bacterial phytochromes for near-infrared imaging, sensing, and light-control in mammals. Chem. Soc. Rev. 42, 3441–3452 (2013).
Shcherbakova, D. M. Near-infrared and far-red genetically encoded indicators of neuronal activity. J. Neurosci. Meth. 362, 109314 (2021).
Manoilov, K. Y., Verkhusha, V. V. & Shcherbakova, D. M. A guide to the optogenetic regulation of endogenous molecules. Nat. Methods 18, 1027–1037 (2021).
Kaberniuk, A. A., Baloban, M., Monakhov, M. V., Shcherbakova, D. M. & Verkhusha, V. V. Single-component near-infrared optogenetic systems for gene transcription regulation. Nat. Commun. 12, 3859 (2021).
Reichhart, E., Ingles-Prieto, A., Tichy, A. M., McKenzie, C. & Janovjak, H. A phytochrome sensory domain permits receptor activation by red light. Angew. Chem. Int. Ed. Engl. 55, 6339–6342 (2016).
Arkhipov, A. et al. Architecture and membrane interactions of the EGF receptor. Cell 152, 557–569 (2013).
Maruyama, I. N. Mechanisms of activation of receptor tyrosine kinases: monomers or dimers. Cells 3, 304–330 (2014).
Shen, J. et al. Extracellular juxtamembrane motif critical for TrkB preformed dimer and activation. Cells 8, 932 (2019).
Sparrow, L. G. et al. The disulfide bonds in the C-terminal domains of the human insulin receptor ectodomain. J. Biol. Chem. 272, 29460–29467 (1997).
Bell, C. A. et al. Rotational coupling of the transmembrane and kinase domains of the Neu receptor tyrosine kinase. Mol. Biol. Cell 11, 3589–3599 (2000).
Leopold, A. V., Pletnev, S. & Verkhusha, V. V. Bacterial phytochrome as a scaffold for engineering of receptor tyrosine kinases controlled with near-infrared light. J. Mol. Biol. 432, 3749–3760 (2020).
Leopold, A. V., Chernov, K. G., Shemetov, A. A. & Verkhusha, V. V. Neurotrophin receptor tyrosine kinases regulated with near-infrared light. Nat. Commun. 10, 1129 (2019).
Takala, H., Lehtivuori, H., Hammaren, H., Hytonen, V. P. & Ihalainen, J. A. Connection between absorption properties and conformational changes in Deinococcus radiodurans phytochrome. Biochemistry 53, 7076–7085 (2014).
Burgie, E. S., Zhang, J. & Vierstra, R. D. Crystal structure of Deinococcus phytochrome in the photoactivated state reveals a cascade of structural rearrangements during photoconversion. Structure 24, 448–457 (2016).
Bjorling, A. et al. Structural photoactivation of a full-length bacterial phytochrome. Sci. Adv. 2, e1600920 (2016).
Gotoh, N. Regulation of growth factor signaling by FRS2 family docking/scaffold adaptor proteins. Cancer Sci. 99, 1319–1325 (2008).
Gasser, C. et al. Engineering of a red-light-activated human cAMP/cGMP-specific phosphodiesterase. Proc. Natl Acad. Sci. USA 111, 8803–8808 (2014).
Gupta, V. K., You, Y., Gupta, V. B., Klistorner, A. & Graham, S. L. TrkB receptor signalling: implications in neurodegenerative, psychiatric and proliferative disorders. Int. J. Mol. Sci. 14, 10122–10142 (2013).
Honarvar, H. et al. Evaluation of HER2-specific peptide ligand for its employment as radiolabeled imaging probe. Sci. Rep. 8, 2998 (2018).
Singh, B., Carpenter, G. & Coffey, R. J. EGF receptor ligands: recent advances. F1000Res. 5, 2270 (2016).
Gutierrez, C. & Schiff, R. HER2: biology, detection, and clinical implications. Arch. Pathol. Lab. Med. 135, 55–62 (2011).
D’Uva, G. et al. ERBB2 triggers mammalian heart regeneration by promoting cardiomyocyte dedifferentiation and proliferation. Nat. Cell Biol. 17, 627–638 (2015).
Barash, S., Wang, W. & Shi, Y. Human secretory signal peptide description by hidden Markov model and generation of a strong artificial signal peptide for secreted protein expression. Biochem. Biophys. Res. Commun. 294, 835–842 (2002).
Barlowe, C. Signals for COPII-dependent export from the ER: what’s the ticket out? Trends Cell Biol. 13, 295–300 (2003).
Ma, D. et al. Golgi export of the Kir2.1 channel is driven by a trafficking signal located within its tertiary structure. Cell 145, 1102–1115 (2011).
Lu, P. et al. Accurate computational design of multipass transmembrane proteins. Science 359, 1042–1046 (2018).
Joo, D. et al. Biphasic activation of extracellular signal-regulated kinase (ERK) 1/2 in epidermal growth factor (EGF)-stimulated SW480 colorectal cancer cells. BMB Rep. 49, 220–225 (2016).
Huang, Z. et al. Two FGF receptor kinase molecules act in concert to recruit and transphosphorylate phospholipase cgamma. Mol. Cell 61, 98–110 (2016).
Chen, T. W. et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300 (2013).
Minichiello, L. TrkB signalling pathways in LTP and learning. Nat. Rev. Neurosci. 10, 850–860 (2009).
Bjorkholm, C. & Monteggia, L. M. BDNF—a key transducer of antidepressant effects. Neuropharmacology 102, 72–79 (2016).
Faraguna, U., Vyazovskiy, V. V., Nelson, A. B., Tononi, G. & Cirelli, C. A causal role for brain-derived neurotrophic factor in the homeostatic regulation of sleep. J. Neurosci. 28, 4088–4095 (2008).
Huber, R., Tononi, G. & Cirelli, C. Exploratory behavior, cortical BDNF expression, and sleep homeostasis. Sleep 30, 129–139 (2007).
Hairston, I. S. et al. Sleep deprivation effects on growth factor expression in neonatal rats: a potential role for BDNF in the mediation of delta power. J. Neurophysiol. 91, 1586–1595 (2004).
Taishi, P. et al. Conditions that affect sleep alter the expression of molecules associated with synaptic plasticity. Am. J. Physiol. Regul. Integr. Comp. Physiol. 281, R839–R845 (2001).
Kushikata, T., Fang, J. & Krueger, J. M. Brain-derived neurotrophic factor enhances spontaneous sleep in rats and rabbits. Am. J. Physiol. 276, R1334–R1338 (1999).
Bachmann, V. et al. The BDNF Val66Met polymorphism modulates sleep intensity: EEG frequency- and state-specificity. Sleep 35, 335–344 (2012).
Uda, Y. et al. Efficient synthesis of phycocyanobilin in mammalian cells for optogenetic control of cell signaling. Proc. Natl Acad. Sci. USA 114, 11962–11967 (2017).
Dehkhoda, F., Lee, C. M. M., Medina, J. & Brooks, A. J. The growth hormone receptor: mechanism of receptor activation, cell signaling, and physiological aspects. Front Endocrinol. 9, 35 (2018).
Watowich, S. S. The erythropoietin receptor: molecular structure and hematopoietic signaling pathways. J. Investig. Med 59, 1067–1072 (2011).
Maghazachi, A. A. Insights into seven and single transmembrane-spanning domain receptors and their signaling pathways in human natural killer cells. Pharm. Rev. 57, 339–357 (2005).
Botos, I., Segal, D. M. & Davies, D. R. The structural biology of Toll-like receptors. Structure 19, 447–459 (2011).
Fleishman, S. J., Harrington, S., Friesner, R. A., Honig, B. & Ben-Tal, N. An automatic method for predicting transmembrane protein structures using cryo-EM and evolutionary data. Biophys. J. 87, 3448–3459 (2004).
Unterreitmeier, S. et al. Phenylalanine promotes interaction of transmembrane domains via GxxxG motifs. J. Mol. Biol. 374, 705–718 (2007).
Bathina, S. & Das, U. N. Brain-derived neurotrophic factor and its clinical implications. Arch. Med Sci. 11, 1164–1178 (2015).
Kraemer, B. R., Yoon, S. O. & Carter, B. D. The biological functions and signaling mechanisms of the p75 neurotrophin receptor. Handb. Exp. Pharmacol. 220, 121–164 (2014).
Numakawa, T. & Odaka, H. Brain-derived neurotrophic factor signaling in the pathophysiology of Alzheimer’s disease: beneficial effects of flavonoids for neuroprotection. Int. J. Mol. Sci. 22, 5719 (2021).
Sungur, A. O. et al. Aberrant cognitive phenotypes and altered hippocampal BDNF expression related to epigenetic modifications in mice lacking the post-synaptic scaffolding protein SHANK1: implications for autism spectrum disorder. Hippocampus 27, 906–919 (2017).
Castren, E. & Monteggia, L. M. Brain-derived neurotrophic factor signaling in depression and antidepressant action. Biol. Psychiatry 90, 128–136 (2021).
Zielinski, M. R., Gerashchenko, L., Karpova, S. A. & Gerashchenko, D. A novel telemetric system to measure polysomnographic biopotentials in freely moving animals. J. Neurosci. Methods 216, 79–86 (2013).
Morairty, S. R. et al. A role for cortical nNOS/NK1 neurons in coupling homeostatic sleep drive to EEG slow wave activity. Proc. Natl Acad. Sci. USA 110, 20272–20277 (2013).
Prerau, M. J., Brown, R. E., Bianchi, M. T., Ellenbogen, J. M. & Purdon, P. L. Sleep neurophysiological dynamics through the lens of multitaper spectral analysis. Physiol. 32, 60–92 (2017).
Acknowledgements
We thank J. Ihalainen (University of Jyväskylä, Finland) for the DrBphP gene and D. Lindholm (University of Helsinki, Finland) for the cell lines. This work was supported by the grants GM122567, AG061774 and NS106406 from the US National Institutes of Health and 322226 from the Academy of Finland.
Author information
Authors and Affiliations
Contributions
A.V.L. engineered and characterized the optogenetic constructs. S.T. and C.Y. performed animal and histochemical studies. V.V.V. planned and directed the project and together with A.V.L. and D.G. designed the experiments, analyzed the data and wrote the manuscript. All authors revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Methods thanks Bianxiao Cui, Benjamin Rost and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Rita Strack was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Structural predeterminants of opto-RTK engineering.
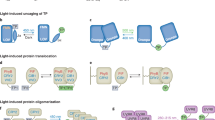
(a) Various hypotheses of RTK activation. Top: RTK dimerization: inactive RTKs exist as monomers and ligand binding causes their dimerization and consequent activation. Bottom: Rotational coupling: inactive RTKs exist as preformed dimers and ligand binding causes conformational changes resulting in RTK activation. (b) Structural changes occurring in EGFR receptor. Left: EGFR receptor exists as a preformed inactive dimer. Right: EGF ligand binding causes conformational changes and EGFR activation. Ab initio models of EGFR in active and inactive states are modified with permission from15. (c) Light-induced conformational changes in DrBphP–PCM obligate dimer cause distance increase between C-termini of DrBphP-PCM protomers17. (d) Schematic representation of proposed opto-RTK design (top) and mechanism of its activation (bottom). DrBphP-PCM targeted to the extracellular surface by Igκ signaling peptide is connected to cytoplasmic RTK domain (cytoRTK) via transmembrane (tmRTK) domain. In darkness or near-infrared light, opto-RTK remains inactive. Far-red light causes DrBphP-PCM conformational changes, which are transmitted to cytoplasmic RTK domains, causing their re-orientation and trans-phosphorylation.
Extended Data Fig. 2 Engineering and characterization of opto-EGFR and opto-HER2 prototypes.
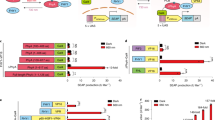
(a) Left: Ligand binding causes RTK autophosphorylation, interaction with Grb2 and SOS, and results in activation of ERK1/2 pathway consisting of RAS, RAF, MEK, and ERK1/2 kinases. Consequently, ERK1/2 activation leads to immediate early gene (IEG) expression driven by transcription factor Elk-1. Right: Likewise, activation of chimeric opto-RTK with far-red light leads to activation of ERK1/2 pathway and induction of Elk-1 dependent IEG expression. (b) Scheme of luciferase reporter assay. Top: ERK1/2 is inactive, and Elk-1 fused to Gal4DBD is monomeric and inactive. Bottom: ERK1/2 is active, and activated ERK1/2 phosphorylates Elk-1. Phosphorylated Elk-1-Gal4DBD fusion dimerizes, binds to 5x UAS sequence, and drives Firefly luciferase (FLuc) reporter expression. (c, d) Light-induced activation of Elk-1-dependent Firefly luciferase reporter expression by opto-EGFR (c) and opto-HER2 (d) prototypes in PC6-3 cells. Renilla luciferase (RLuc) was co-transfected as a normalization control. In the darkness, Firefly luciferase expression is suppressed. 660 nm light activates eDrRTKs and upregulates Firefly luciferase expression. Expression of Firefly and Renilla luciferases was analyzed after 24 h of illumination. Relative luciferase expression activity (RLA) is shown as a ratio of the Firefly luciferase expression relative bioluminescence units to the Renilla luciferase expression relative bioluminescence units (FLuc/RLuc). 25 µM BV was added to the culture medium in all experiments. Data are presented as mean values, error bars represent standard deviation (s.d.), (n≥3, transfection experiments).
Extended Data Fig. 3 Regulation of PLCγ signaling by eDrRTKs.
(a) Top: eDrRTK activates PLCγ. PLCγ catalyzes PIP2 hydrolysis and formation of IP3 and DAG. IP3 interacts with IP3R channels in ER after which they become permeable to Ca2+. In turn, Ca2+ interacts with ORAI channels in the plasma membrane and induces Ca2+ entry from the extracellular space. Bottom: Scheme of the plasmid encoding eDrRTK and GCaMP6m Ca2+ indicator via IRES2. (b) Western blots of phospho-PLCγ, total PLCγ and GAPDH in HEK293 cell lysates and quantification of lane intensities of phospho-PLCγ normalized to GAPDH lane intensities (LIs). (c) Representative HEK293 cells co-transfected with eDrRTKs and GCaMP6m imaged before, during, and after 25 s illumination with 660 nm FR light, and their corresponding calcium flux traces are shown. Red arrows indicate the time when 660 nm light-activation started. After 25 s of 660 nm illumination, inactivating 780 nm light was switched on. Black arrows 1, 2 and 3 indicate time-points when confocal images before illumination, at 660 nm illumination, and after illumination were taken, respectively. 25 µM BV was added to the culture medium in all experiments. Scale bars, 10 µm. Data are presented as mean values, error bars represent standard deviation (s.d.), (n≥3, transfection experiments).
Supplementary information
Supplementary Information
Supplementary Figs. 1–21, Table 1 and References.
Supplementary Data
A workbook with tabs, containing statistical source data for the Supplementary Information.
Source data
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Unprocessed western blots and statistical source data.
Source Data Fig. 4
Unprocessed western blots and statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Unprocessed western blots and statistical source data.
Rights and permissions
About this article
Cite this article
Leopold, A.V., Thankachan, S., Yang, C. et al. A general approach for engineering RTKs optically controlled with far-red light. Nat Methods 19, 871–880 (2022). https://doi.org/10.1038/s41592-022-01517-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-022-01517-z