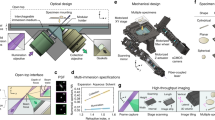
Abstract
Light-sheet microscopy has emerged as the preferred means for high-throughput volumetric imaging of cleared tissues. However, there is a need for a flexible system that can address imaging applications with varied requirements in terms of resolution, sample size, tissue-clearing protocol, and transparent sample-holder material. Here, we present a ‘hybrid’ system that combines a unique non-orthogonal dual-objective and conventional (orthogonal) open-top light-sheet (OTLS) architecture for versatile multi-scale volumetric imaging. We demonstrate efficient screening and targeted sub-micrometer imaging of sparse axons within an intact, cleared mouse brain. The same system enables high-throughput automated imaging of multiple specimens, as spotlighted by a quantitative multi-scale analysis of brain metastases. Compared with existing academic and commercial light-sheet microscopy systems, our hybrid OTLS system provides a unique combination of versatility and performance necessary to satisfy the diverse requirements of a growing number of cleared-tissue imaging applications.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The customized ZEMAX files, CAD files, list of components, and a summary of all datasets and the associated imaging parameters are available as Supplementary Data. Due to the large size of the imaging datasets collected within this manuscript, the datasets are not available in a public repository and are available from the authors upon request.
Code availability
The simulation codes used to model the lateral and axial resolution of the various microscope architectures is available on GitHub. The acquisition software code for the microscope is available from the authors upon request.
References
Chung, K. et al. Structural and molecular interrogation of intact biological systems. Nature 497, 332–337 (2013).
Tanaka, N. et al. Whole-tissue biopsy phenotyping of three-dimensional tumours reveals patterns of cancer heterogeneity. Nat. Biomed. Eng. 1, 796 (2017).
Richardson, D. S. & Lichtman, J. W. Clarifying tissue clearing. Cell 162, 246–257 (2015).
Pan, C. et al. Shrinkage-mediated imaging of entire organs and organisms using uDISCO. Nat. Methods 13, 859–867 (2016).
Renier, N. et al. iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell 159, 896–910 (2014).
Tainaka, K. et al. Chemical landscape for tissue clearing based on hydrophilic reagents. Cell Rep. 24, 2196–2210.e9 (2018).
Susaki, E. A. et al. Advanced CUBIC protocols for whole-brain and whole-body clearing and imaging. Nat. Protoc. 10, 1709–1727 (2015).
Chung, K. & Deisseroth, K. CLARITY for mapping the nervous system. Nat. Methods 10, 508–513 (2013).
Susaki, E. A. et al. Versatile whole-organ/body staining and imaging based on electrolyte-gel properties of biological tissues. Nat. Commun. 11, 1982 (2020).
Jing, D. et al. Tissue clearing of both hard and soft tissue organs with the PEGASOS method. Cell Res. 28, 803–818 (2018).
Power, R. M. & Huisken, J. A guide to light-sheet fluorescence microscopy for multiscale imaging. Nat. Methods 14, 360 (2017).
Huisken, J. & Stainier, D. Y. R. Selective plane illumination microscopy techniques in developmental biology Development 136, 1963–1975 (2009).
Dodt, H.-U. et al. Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain. Nat. Methods 4, 331–336 (2007).
Migliori, B. et al. Light sheet theta microscopy for rapid high-resolution imaging of large biological samples. BMC Biol. 16, 57 (2018).
Tomer, R. et al. Advanced CLARITY for rapid and high-resolution imaging of intact tissues. Nat. Protoc. 9, 1682–1697 (2014).
Voigt, F. F. et al. The mesoSPIM initiative: open-source light-sheet microscopes for imaging cleared tissue. Nat. Methods 16, 1105–1108 (2019).
Chakraborty, T. et al. Light-sheet microscopy of cleared tissues with isotropic, subcellular resolution. Nat. Methods 16, 1109–1113 (2019).
Chen, Y. et al. A versatile tiling light sheet microscope for imaging of cleared tissues. Cell Rep. 33, 108349 (2020).
Kumar, A. et al. Dual-view plane illumination microscopy for rapid and spatially isotropic imaging. Nat. Protoc. 9, 2555–2573 (2014).
Strnad, P. et al. Inverted light-sheet microscope for imaging mouse pre-implantation development. Nat. Methods 13, 139–142 (2015).
McGorty, R. et al. Open-top selective plane illumination microscope for conventionally mounted specimens. Opt. Express 23, 16142–16153 (2015).
McGorty, R., Xie, D. & Huang, B. High-NA open-top selective-plane illumination microscopy for biological imaging. Opt. Express 25, 17798–17810 (2017).
Glaser, A. K. et al. Light-sheet microscopy for slide-free non-destructive pathology of large clinical specimens. Nat. Biomed. Eng. 1, 0084 (2017).
Glaser, A. K. et al. Multi-immersion open-top light-sheet microscope for high-throughput imaging of cleared tissues. Nat. Commun. 10, 1–8 (2019).
Barner, L. A. et al. Solid immersion meniscus lens (SIMlens) for open-top light-sheet microscopy. Opt. Lett. 44, 4451–4454 (2019).
Barner, L. A. et al. Multi-resolution open-top light-sheet microscopy to enable efficient 3D pathology workflows. Biomed. Opt. Express 11, 6605–6619 (2020).
Botcherby, E. J. et al. An optical technique for remote focusing in microscopy. Opt. Commun. 281, 880–887 (2008).
Dunsby, C. Optically sectioned imaging by oblique plane microscopy. Opt. Express 16, 20306–20316 (2008).
Voleti, V. et al. Real-time volumetric microscopy of in vivo dynamics and large-scale samples with SCAPE 2.0. Nat. Methods 16, 1054–1062 (2019).
Bouchard, M. B. et al. Swept confocally-aligned planar excitation (SCAPE) microscopy for high-speed volumetric imaging of behaving organisms. Nat. Photonics 9, 113–119 (2015).
Yang, B. et al. Epi-illumination SPIM for volumetric imaging with high spatial-temporal resolution. Nat. Methods 16, 501–504 (2019).
Millett-Sikking, A. et al. High NA single-objective light-sheet. https://andrewgyork.github.io/high_na_single_objective_lightsheet/index.html (2019).
Kumar, M. et al. Integrated one- and two-photon scanned oblique plane illumination (SOPi) microscopy for rapid volumetric imaging. Opt. Express 26, 13027–13041 (2018).
Hoffmann, M. & Judkewitz, B. Diffractive oblique plane microscopy. Optica 6, 5 (2019).
Sapoznik, E. et al. A versatile oblique plane microscope for large-scale and high-resolution imaging of subcellular dynamics. eLife 9, e57681 (2020).
Yang, B. et al. DaXi-high-resolution, large imaging volume and multi-view single-objective light-sheet microscopy. Nat. Methods 19, 461–469 (2022).
Li, T. et al. Axial plane optical microscopy. Sci. Rep. 4, 7253 (2014).
Kim, J. et al. Oblique-plane single-molecule localization microscopy for tissues and small intact animals. Nat. Methods 16, 853–857 (2019).
Bishop, K. W., Glaser, A. K. & Liu, J. T. C. Performance tradeoffs for single- and dual-objective open-top light-sheet microscope designs: a simulation-based analysis. Biomed. Opt. Express 11, 4627–4650 (2020).
Economo, M. N. et al. A platform for brain-wide imaging and reconstruction of individual neurons. eLife 5, e10566 (2016).
Winnubst, J. et al. Reconstruction of 1,000 projection neurons reveals new cell types and organization of long-range connectivity in the mouse brain. Cell 179, 268–281.e13 (2019).
Kubota, S. I. et al. Whole-body profiling of cancer metastasis with single-cell resolution. Cell Rep. 20, 236–250 (2017).
Planchon, T. A. et al. Rapid three-dimensional isotropic imaging of living cells using Bessel beam plane illumination. Nat. Methods 8, 417–423 (2011).
Dean, K. et al. Deconvolution-free subcellular imaging with axially swept light sheet microscopy. Biophys. J. 108, 2807–2815 (2015).
Sparks, H. et al. Dual-view oblique plane microscopy (dOPM). Biomed. Opt. Express 11, 7204–7220 (2020).
Keller, P. J. et al. Fast, high-contrast imaging of animal development with scanned light sheet-based structured-illumination microscopy. Nat. Methods 7, 637–642 (2010).
Yueqian, Z. & Herbert, G. Systematic design of microscope objectives. Part I: system review and analysis. Adv. Opt. Technol. 8, 313–347 (2019).
Hörl, D. et al. BigStitcher: reconstructing high-resolution image datasets of cleared and expanded samples. Nat. Methods 16, 870–874 (2019).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Balazs, B., et al. A real-time compression library for microscopy images. Preprint at bioRxiv https://doi.org/10.1101/164624 (2017).
Chan, K. Y. et al. Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat. Neurosci. 20, 1172–1179 (2017).
Chi, J. et al. Three-dimensional adipose tissue imaging reveals regional variation in beige fat biogenesis and PRDM16-dependent sympathetic neurite density. Cell Metab. 27, 226–236.e3 (2018).
Ehata, S. et al. Transforming growth factor-β promotes survival of mammary carcinoma cells through induction of antiapoptotic transcription factor DEC1. Cancer Res. 67, 9694 (2007).
Nishida, J. et al. Epigenetic remodelling shapes inflammatory renal cancer and neutrophil-dependent metastasis. Nat. Cell Biol. 22, 465–475 (2020).
Miyoshi, H., Takahashi, M., Gage, F. H. & Verma, I. M. Stable and efficient gene transfer into the retina using an HIV-based lentiviral vector. Proc. Natl Acad. Sci. USA 94, 10319–10323 (1997).
Matsumoto, K. et al. Advanced CUBIC tissue clearing for whole-organ cell profiling. Nat. Protoc. 14, 3506–3537 (2019).
Acknowledgements
We would like to thank A. York and A. Millet-Sikking for discussions regarding oblique planar microscopy, remote focus imaging, and alignment procedures. We would also like to thank J. Daniels for discussions and his development of the multi-immersion objective, C. Shimizu (RIKEN BDR) for the support of preparing CUBIC-cleared and stained specimens, and K. Miyazono (The University of Tokyo) for the discussion and support of brain metastasis experiments. This work was funded in part by the National Institutes of Health (NIH) K99 CA240681 (A.K.G.), R01CA244170 (J.T.C.L.), R01EB031002 (J.T.C.L.), R01GM079712 (T.I.), R01DK107436 (L.X.), R01DK092202 (L.X.), R01MH117820 and R01NS104949 (R.C.R.); Department of Defense (DoD) Prostate Cancer Research Program (PCRP) W81XWH-18-10358 (J.T.C.L. and L.D.T.), W81XWH-19-1-0589 (N.P.R.), W81XWH-20-1-0039 (X.W.), and Prostate Cancer Young Investigator Award (N.P.R.); National Science Foundation (NSF) Graduate Research Fellowship DGE-1762114 (L.A.B. and K.W.B.); NSF 1934292 HDR: I-DIRSE-FW (J.T.C.L. and R.B.S.); Washington Research Foundation Postdoctoral Fellowship (C.R.S.); Science and Technology Platform Program for Advanced Biological Medicine by the Japan Agency for Medical Research and Development (AMED) JP21am0401011 (H.R.U.), ERATO by Japan Science and Technology Agency (JST) JPMJER2001 (H.R.U.), HFSP Research Grant Program RGP0019/2018 (H.R.U.), AMED-PRIME JP21gm6210027 (E.A.S.), Grants-in-Aid for Scientific Research on Innovative Areas (JSPS KAKENHI grant) 17H06328 (E.A.S.), Grants-in-Aid for Scientific Research on Innovative Areas (JSPS KAKENHI grant) 20K1612 (S.I.K.). Work in the Murawala laboratory is supported by grants from NIH-COBRE (5P20GM104318-08) and DFG (429469366). Work in the Dodt laboratory is supported by grants FWF P31263-B26 and WWTF CS19-019.
Author information
Authors and Affiliations
Contributions
A.K.G. and J.T.C.L. conceived of and designed the microscope system. P.R.N. provided feedback on the system design and its potential applications. A.K. G., K.W.B., R.B.S., and G.G. performed simulations of the microscope. A.K.G. fabricated the microscope system with help from L.A.B.E.A.S. and H.R.U. provided and prepared the immunostained CUBIC-cleared mouse brains. E.A.S., S.I.K., and H.R.U. prepared and provided the metastatic mouse brains. J.C. and K.S. prepared and provided the mouse brain. P.B., E.T., and R.C.R. provided the mouse brain with preparation by A.K.G. The human brain slice was prepared by A.K.G.H.L. and L.A.G.L. provided quantitative analysis of the high-resolution images. Y.Y. and H.Z. provided and prepared the PEGASOS-cleared mouse brain. E.K.N., B.J.B., and J.S. provided and prepared the SHIELD-cleared mouse embryos. H.H., N.P.R., and L.D.T. provided and prepared the ECi-cleared human prostate tissue. J.J.W., R.S., E.S., C.R.S., and M.Y.G. provided and prepared the Ce3D-cleared mouse lymph node. X.W. and L.X. provided and prepared the iDISCO-cleared mouse prostate. A.K.H. and T.I. provided and prepared the ClearSee-processed Arabidopsis plant and M.P., P.M., and H.U.D. provided and prepared the DEEP-Clear-processed Axolotl. A.K.G. and J.T.C.L. led the writing of the manuscript. All authors contributed to the manuscript.
Corresponding authors
Ethics declarations
Competing interests
A.K.G., N.P.R., L.D.T., and J.T.C.L. are co-founders and shareholders of Alpenglow Biosciences. L.A.G.L. and H.L. are employees of Leica Microsystems, maker of the Aivia software.
Peer review
Peer review information
Nature Methods thanks Peter Santi, Per Uhlen, and Fabian Voigt for their contribution to the peer review of this work. Rita Strack was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Regions of interests from metastatic brain specimens.
All n = 34 metastatic colonies collected from the metastatic brain specimens (3 brains from OS-RC-2 cancer cell line, 3 brains from MDA-MB-231 cancer cell line). All scale bars represent 10 μm.
Extended Data Fig. 2 Multi-scale 3D pathology of human prostate tissue.
(a) Fast meso-scale screening results for a multi-cm-sized piece of human prostate tissue stained with TO-PRO-3 Iodide (nuclear) and Eosin. Representative high-resolution regions of interests for two different foci of cancer are shown in (b) and (c). The insets demonstrate the ability to clearly resolve sub-nuclear features in cancer nuclei. Scale bar lengths are as follows: (a) 1 cm, (b) and (c) 10 μm. The imaging data in (a-c) was acquired from in a single experiment, which was not repeated.
Extended Data Fig. 3 Assessment of 3D cell proliferation assays with iDISCO.
(a) Screening of an intact mouse prostate cleared using iDISCO, labeled with TOPRO3 Iodide (nuclear) and an EduClick cell-proliferation marker. A higher-resolution region of interest focused on a prostate gland is shown in (b). An additional zoomed-in view showing the ability to resolve individual proliferating nuclei (c). Scale bars lengths are as follows: (a) 1 mm, (b) 200 μm, and (c) 100 μm. The imaging data in (a-c) was acquired from in a single experiment, which was not repeated.
Extended Data Fig. 4 Imaging of non-rodent and non-human tissues.
(a) Hybrid open-top light-sheet imaging of a ClearSee-cleared Arabidopsis specimen. (b) Higher-resolution imaging of the Arabidopsis root. (c) Meso-scale imaging of a large multi-cm Axolotl cleared with DEEP-Clear. Scale bars lengths are as follows: (a) 1 mm, (b) 100 μm, and (c) 1 cm. The imaging data in (a-c) was acquired from in a single experiment, which was not repeated.
Extended Data Fig. 5 Imaging of immunostained and endogenously fluorescent CUBIC-cleared mouse brains.
Hybrid OTLS imaging of whole mouse brains. (a-c) with endogenously preserved GCaMP6s fluorescence, and immunostained with (d) anti-ChAT antibody + SYTOX-G or (e) anti-Parvalbumin (PV) antibody + SYTOX-G by CUBIC-HistoVision. Scale bar lengths are as follows: (a-b) 1 mm, (c) 10 μm, (d-e) 2 mm. The imaging data in (a-e) was acquired from in a single experiment, which was not repeated.
Extended Data Fig. 6 Large-scale imaging of thick human tissues.
(a) Slab of human brain tissue after CUBIC clearing. A mouse brain is shown for size comparison. (b) ODO imaging results of the entire 3-mm thick brain slab. Autofluorescence is shown in black and white, and the amyloid small molecule stain (pFTAA) is shown in green. (c) Zoom-in views of an amyloid-rich region reveal perivascular accumulation. Scale bar lengths are as follows: (b) 1 cm. The imaging data in (a-c) was acquired from in a single experiment, which was not repeated.
Extended Data Fig. 7 NODO imaging quality versus imaging depth in a CUBIC-cleared mouse brain.
(a) Multiple two-channel regions of interest (ROIs) were acquired from a CUBIC-cleared mouse brain at various imaging depths (z = 1–8 mm). (b-e) xy views for ROIs at various depths are shown. The corresponding xz views of each ROI are shown in (f-i). The high-magnification insets show fine sub-nuclear details and demonstrate that there is minimal degradation in image quality as a function of depth. All scalebars denote 10 µm. The imaging data in (b-i) was acquired from in a single experiment, which was not repeated.
Extended Data Fig. 8 NODO imaging quality versus imaging depth in a PEGASOS-cleared mouse brain.
(a) A single ROI was acquired from a PEGASOS-cleared mouse brain. (b) xz view of the ~5 mm deep ROI, demonstrating near-consistent image quality versus depth. (c-g) xy views for ROIs at various depths are shown. The high-magnification insets show fine sub-nuclear details and demonstrate that there is minimal degradation in image quality as a function of depth. Scale bar lengths are as follows: (b) 100 µm, (c-g) 10 µm. The imaging data in (b-g) was acquired from in a single experiment, which was not repeated.
Extended Data Fig. 9 NODO imaging quality versus imaging depth in an ECi-cleared mouse brain.
(a) Multiple regions of interest (ROIs) were acquired from an ECi-cleared mouse brain at various imaging depths (z = 0–5 mm). (b-f) xy views for ROIs at various depths are shown. The corresponding xz views of each ROI are shown in (g-k). The high-magnification insets show fine sub-nuclear details and demonstrate that there is minimal degradation in image quality as a function of depth. All scalebars denote 10 µm. The imaging data in (b-k) was acquired from in a single experiment, which was not repeated.
Supplementary information
Supplementary Information
Supplementary Notes 1–6, Supplementary Methods, Supplementary Tables 1–3, Supplementary Figures 1–25, References
Supplementary Video 1
Live video showing assembly and operation of the hybrid open-top light-sheet microscope.
Supplementary Video 2
ODO and NODO imaging results from a CUBIC-cleared mouse brain stained with SYTOX Green and αSMA.
Supplementary Video 3
Sparse axonal imaging results from an ECI-cleared Slc17a7-Cre mouse brain.
Supplementary Video 4
Multi-scale imaging of MDA-MB-231 and OS-RC-2 metastatic lesions in CUBIC-cleared mouse brains.
Supplementary Video 5
ODO and NODO imaging results from a PEGASOS-cleared Thy1-EGFP mouse brain.
Supplementary Video 6
Multi-channel 3D imaging of a Ce3D-cleared mouse lymph node.
Supplementary Video 7
Assessment of 3D cell proliferation in an iDISCO-cleared mouse prostate.
Supplementary Video 8
ODO imaging results from CUBIC-HV cleared and stained mouse brains.
Supplementary Video 9
Imaging results from a CUBIC-cleared Slc17a7-IRES2-Cre;Ai14 mouse brain.
Supplementary Video 10
Imaging results from a CUBIC-cleared Chat-IRES-Cre;Ai162 mouse brain.
Supplementary Video 11
Large-scale imaging of a thick CUBIC-cleared human brain slab.
Supplementary Data
ZEMAX files for the hybrid open-top light-sheet microscope
Supplementary Data
CAD files for the hybrid open-top light-sheet microscope
Supplementary Data
Parts list for the hybrid open-top light-sheet microscope
Supplementary Data
Summary of experimental imaging parameters for datasets
Supplementary Data
Specifications of existing immersion objective lenses
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Glaser, A.K., Bishop, K.W., Barner, L.A. et al. A hybrid open-top light-sheet microscope for versatile multi-scale imaging of cleared tissues. Nat Methods 19, 613–619 (2022). https://doi.org/10.1038/s41592-022-01468-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-022-01468-5
This article is cited by
-
Super-sectioning with multi-sheet reversible saturable optical fluorescence transitions (RESOLFT) microscopy
Nature Methods (2024)
-
Signal improved ultra-fast light-sheet microscope for large tissue imaging
Communications Engineering (2024)
-
S-polarized light-sheets improve resolution and light-efficiency in oblique plane microscopy
Scientific Reports (2024)
-
An end-to-end workflow for nondestructive 3D pathology
Nature Protocols (2024)
-
Benchtop mesoSPIM: a next-generation open-source light-sheet microscope for cleared samples
Nature Communications (2024)