Abstract
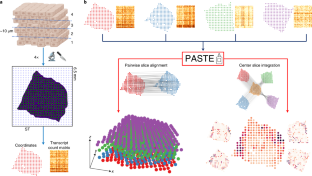
Spatial transcriptomics (ST) measures mRNA expression across thousands of spots from a tissue slice while recording the two-dimensional (2D) coordinates of each spot. We introduce probabilistic alignment of ST experiments (PASTE), a method to align and integrate ST data from multiple adjacent tissue slices. PASTE computes pairwise alignments of slices using an optimal transport formulation that models both transcriptional similarity and physical distances between spots. PASTE further combines pairwise alignments to construct a stacked 3D alignment of a tissue. Alternatively, PASTE can integrate multiple ST slices into a single consensus slice. We show that PASTE accurately aligns spots across adjacent slices in both simulated and real ST data, demonstrating the advantages of using both transcriptional similarity and spatial information. We further show that the PASTE integrated slice improves the identification of cell types and differentially expressed genes compared with existing approaches that either analyze single ST slices or ignore spatial information.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Code availability
The PASTE methods are implemented in an open-source, publicly available Python package that is available at https://github.com/raphael-group/paste. All the code to reproduce the analysis can be found at https://github.com/raphael-group/paste_reproducibility.
References
Ståhl, P. L. et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353, 78–82 (2016).
10x Genomics. Visium spatial gene expression: map the whole transcriptome within the tissue context. https://www.10xgenomics.com/products/spatial-gene-expression/ (accessed October 2020) (2019).
Zhao, E. et al. Spatial transcriptomics at subspot resolution with bayesspace.Nat. Biotechnol. 39, 1375–1384 (2021).
Berglund, E. et al. Spatial maps of prostate cancer transcriptomes reveal an unexplored landscape of heterogeneity. Nat. Commun. 9, 2419 (2018).
Thrane, K., Eriksson, H., Maaskola, J., Hansson, J. & Lundeberg, J. Spatially resolved transcriptomics enables dissection of genetic heterogeneity in stage iii cutaneous malignant melanoma. Cancer Res. 78, 5970–5979 (2018).
Moncada, R. et al. Integrating microarray-based spatial transcriptomics and single-cell RNA-seq reveals tissue architecture in pancreatic ductal adenocarcinomas. Nat. Biotechnol. 38, 333–342 (2020).
Ji, A. et al. Multimodal analysis of composition and spatial architecture in human squamous cell carcinoma. Cell 182, 1661–1662 (2020).
Chen, W.-T. et al. Spatial transcriptomics and in situ sequencing to study Alzheimer’s disease. Cell 182, 976–991.e19 (2020).
Lundmark, A. et al. Gene expression profiling of periodontitis-affected gingival tissue by spatial transcriptomics. Sci. Rep. 8, 9370 (2018).
Asp, M. et al. Spatial detection of fetal marker genes expressed at low level in adult human heart tissue. Sci. Rep. 7, 12941 (2017).
Maniatis, S. et al. Spatiotemporal dynamics of molecular pathology in amyotrophic lateral sclerosis. Science 364, 89–93 (2019).
Maynard, K. R. et al. Transcriptome-scale spatial gene expression in the human dorsolateral prefrontal cortex. Nat. Neurosci. 24, 425–436 (2021).
Liu, R. et al. Modeling spatial correlation of transcripts with application to developing pancreas. Sci. Rep. 9, 5592 (2019).
Svensson, V., Teichmann, S. A. & Stegle, O. SpatialDE: identification of spatially variable genes. Nat. Meth. 15, 343 (2018).
Arnol, D., Schapiro, D., Bodenmiller, B., Saez-Rodriguez, J. & Stegle, O. Modeling cell-cell interactions from spatial molecular data with spatial variance component analysis. Cell Rep. 29, 202–211 (2019).
Cang, Z. & Nie, Q. Inferring spatial and signaling relationships between cells from single cell transcriptomic data. Nat. Commun. 11, 2084 (2020).
Ji, N. & Oudenaarden, A. Single-molecule fluorescent in situ hybridization (smFISH) of C. elegans worms and embryos. In WormBook: The Online Review of C. elegans Biology (ed. WormBook) 1–16 (The C. elegans Research Community, 2012).
Eng, C.-H. L. et al. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH+. Nature 568, 235–239 (2019).
Wang, X. et al. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science 361, eaat5691 (2018).
Stickels, R. R. et al. Highly sensitive spatial transcriptomics at near-cellular resolution with slide-seqv2. Nat. Biotechnol. 39, 313–319 (2021).
Elosua-Bayes, M., Nieto, P., Mereu, E., Gut, I. & Heyn, H. SPOTlight: seeded NMF regression to deconvolute spatial transcriptomics spots with single-cell transcriptomes. Nucleic Acids Res. 49, e50–e50 (2021).
Bergenstråhle, J., Larsson, L. & Lundeberg, J. Seamless integration of image and molecular analysis for spatial transcriptomics workflows. BMC Genomics 21, 482 (2020).
Äijö, T. et al. Splotch: robust estimation of aligned spatial temporal gene expression data. Preprint at bioRxiv https://doi.org/10.1101/757096 (2019).
Haghverdi, L., Lun, A. T., Morgan, M. D. & Marioni, J. C. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat. Biotechnol. 36, 421–427 (2018).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019).
Hie, B., Bryson, B. & Berger, B. Efficient integration of heterogeneous single-cell transcriptomes using scanorama. Nat. Biotechnol. 37, 685–691 (2019).
Mandric, I., Hill, B. L., Freund, M. K., Thompson, M. & Halperin, E. Batman: fast and accurate integration of single-cell RNA-seq datasets via minimum-weight matching. iScience 23, 101185 (2020).
Demetci, P., Santorella, R., Sandstede, B., Noble, W. S. & Singh, R. Gromov-Wasserstein optimal transport to align single-cell multi-omics data. Preprint at bioRxiv https://doi.org/10.1101/2020.04.28.066787 (2020).
Tran, H. T. N. et al. A benchmark of batch-effect correction methods for single-cell RNA sequencing data. Genome Biol. 21, 12 (2020).
Titouan, V., Courty, N., Tavenard, R. & Flamary, R. Optimal transport for structured data with application on graphs. In International Conference on Machine Learning (eds Chaudhuri, K. & Salakhutdinov, R.) 6275–6284 (PMLR, 2019).
Lee, D. & Seung, H. Algorithms for non-negative matrix factorization. In Advances in Neural Information Processing Systems 13: Proceedings of the 2000 Conference, NIPS 2000 (Neural Information Processing Systems Foundation, 2001).
Shao, C. & Höfer, T. Robust classification of single-cell transcriptome data by nonnegative matrix factorization. Bioinformatics 33, 235–242 (2016).
Zhu, X., Ching, T., Pan, X., Weissman, S. M. & Garmire, L. Detecting heterogeneity in single-cell RNA-seq data by non-negative matrix factorization. PeerJ 5, e2888 (2017).
Elyanow, R. et al. STARCH: copy number and clone inference from spatial transcriptomics data.Phys. Biol. 18, 035001 (2021).
O’Neill, R. et al. Indices of landscape pattern. Landsc. Ecol. 1, 153–162 (1988).
Maniatis, S. et al. Spatiotemporal dynamics of molecular pathology in amyotrophic lateral sclerosis. Science 364, 89–93 (2019).
Andersson, A. et al. Spatial deconvolution of her2-positive breast cancer delineates tumor-associated cell type interactions. Nat. Commun. 12, 6012 (2021).
Biancalani, T. et al. Deep learning and alignment of spatially resolved single-cell transcriptomes with Tangram.Nat. Methods 18, 1352–1362 (2021).
Yoosuf, N., Navarro, J., Salmén, F., Ståhl, P. L. & Daub, C. O. Identification and transfer of spatial transcriptomics signatures for cancer diagnosis. Breast Cancer Res. 22, 6 (2020).
Brown, L. G. A survey of image registration techniques. ACM Comput. Surv. 24, 325–376 (1992).
Fatras, K., Zine, Y., Flamary, R., Gribonval, R. & Courty, N. Learning with minibatch Wasserstein: asymptotic and gradient properties. In AISTATS, 2131–2141 http://proceedings.mlr.press/v108/fatras20a.html (2020).
Feydy, J. et al. Interpolating between optimal transport and mmd using sinkhorn divergences. In The 22nd International Conference on Artificial Intelligence and Statistics, 2681–2690 (2019).
Marx, V. Method of the year: spatially resolved transcriptomics. Nat. Methods 18, 9–14 (2021).
Larsson, L., Frisén, J. & Lundeberg, J. Spatially resolved transcriptomics adds a new dimension to genomics. Nat. Methods 18, 15–18 (2021).
Wahba, G. A least squares estimate of satellite attitude. SIAM Rev. 7, 409–409 (1965).
Kabsch, W. A solution for the best rotation to relate two sets of vectors. Acta Crystallogr. A 32, 922–923 (1976).
Lin, P., Troup, M. & Ho, J. W. K. Cidr: Ultrafast and accurate clustering through imputation for single-cell RNA-seq data. Genome Biology 18, 59 (2017).
Mongia, A., Sengupta, D. & Majumdar, A. Mcimpute: matrix completion based imputation for single cell RNA-seq data. Frontiers in Genetics 10, 9 (2019).
Hou, W., Ji, Z., Ji, H. & Hicks, S. C. A systematic evaluation of single-cell RNA-sequencing imputation methods. Genome Biology 21, 218 (2020).
Févotte, C. & Cemgil, A. T. Nonnegative matrix factorizations as probabilistic inference in composite models. In 2009 17th European Signal Processing Conference, 1913–1917 (IEEE, 2009).
Durif, G., Modolo, L., Mold, J. E., Lambert-Lacroix, S. & Picard, F. Probabilistic count matrix factorization for single cell expression data analysis. Bioinformatics 35, 4011–4019 (2019).
Townes, F. W., Hicks, S. C., Aryee, M. J. & Irizarry, R. A. Feature selection and dimension reduction for single-cell RNA-seq based on a multinomial model. Genome Biol. 20, 295 (2019).
Elyanow, R., Dumitrascu, B., Engelhardt, B. E. & Raphael, B. J. netnmf-sc: leveraging gene-gene interactions for imputation and dimensionality reduction in single-cell expression analysis. Genome Res.30, 195–204 (2020).
Wolf, F. A., Angerer, P. & Theis, F. J. Scanpy: large-scale single-cell gene expression data analysis. Genome Biology 19, 15 (2018).
Flamary, R. & Courty, N. Pot Python Optimal Transport Library https://pythonot.github.io/ (2017).
Sun, S., Zhu, J., Ma, Y. & Zhou, X. Accuracy, robustness and scalability of dimensionality reduction methods for single-cell RNA-seq analysis. Genome Biol. 20, 269 (2019).
Chen, M. & Zhou, X. Viper: variability-preserving imputation for accurate gene expression recovery in single-cell RNA sequencing studies. Genome Biol. 19, 196 (2018).
Acknowledgements
This work was supported by National Cancer Institute grants U24CA211000 and U24CA248453 to B.J.R. The funder had no role in the conceptualization, design, data collection, analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
R.Z. conceived, designed and developed the method, analyzed the DLPFC and Her2 breast cancer datasets and wrote the manuscript with contributions from the coauthors. M.L. implemented the method and performed the simulation, SCC and spinal cord data analyses. A.S. contributed to the benchmarking of PASTE against Seurat and STUtility and the analyses of the DLPFC and SCC dataset. B.J.R. supervised the work, contributed to the design of the method and wrote the manuscript with contributions from the coauthors. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
B.J.R. is a cofounder of, and consultant to, Medley Genomics. The other authors declare no competing interests.
Peer review
Peer review information
Nature Methods thanks Jean Yang and the other, anonymous, reviewers for their contribution to the peer review of this work. Lei Tang was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 2 PASTE results on simulated data generated from each of the indicated breast cancer slices35.
Each line (color) corresponds to running PASTE with a specific value for alpha. Error bars represent the standard deviation across 10 simulated instances.
Extended Data Fig. 3 Comparison of published clusters and clusters obtained by PASTE on ST data from SCC patients 2, 5, 9, and 10 in21.
(Left) The published cluster labels from21 of spots in slice A from each of the four patients. (Right) k-means clustering of inferred center slice from PASTE.
Extended Data Fig. 4 PASTE integration of Her2 breast cancer patient G from Andersson et al.
(a) Pathological annotations and (b) clustering results from PASTE integrated slice for a slice of breast cancer patient G from Andersson et al. Black circles indicate small region of spots of in situ cancer which are also clustered together in the PASTE integrated slice.
Extended Data Fig. 5 Dorsolateral prefrontal cortex ST data from31.
Each of the three samples is composed of four ST slices. The first two slices and last two slices are 10μm apart while the middle pair of slices is taken 300μm apart. Spots are colored by the six neocortical layers or the white matter according to the annotation of31.
Extended Data Fig. 6 Pairwise alignment of slices B and C from DLPFC Sample I.
Pairwise alignment using (a) PASTE, (b) Seurat, (c) Tangram and (d) STUtility. Gray lines connect the 1000 spot pairs with highest alignment values from each method. PASTE and STUtility alignments are more consistent with spatial organization of slices than Seurat and Tangram alignments.
Extended Data Fig. 7 Alignment accuracy of adjacent DLPFC slices using PASTE with different expression costs.
PASTE with: (Default) All genes and KL divergence, (Lib-Log-Norm) All genes with library size normalization and log transformation and Euclidean distance, (HVG) Same as Lib-Log-Norm but restricted to top 2000 highly variable genes.
Extended Data Fig. 8 TRABD2A expression in a single slice and PASTE integrated slice.
The boundaries between the layers are marked in green in a and c. WM and Layers 6 to 1 have 625, 614, 621, 247, 924, 224 and 380 spots respectively. Inner boxplots show the 25%, 50% and 75% quantiles of the distributions. p-values (rounded to the closest power of 10) for the difference in distribution (two-sided Mann-Whitney U test) between adjacent layers are indicated. TRABD2A was validated using smFISH in31 as a layer 5 marker gene.
Extended Data Fig. 9 Ranking of known layer-specific marker genes by differential expression analysis.
Gene ranking using: the pseudo-bulk approach of Maynard et al., PASTE center slice integration, Scanorama, and Seurat. Red lines indicate median rank of marker genes which are 1147 for Maynard et al, 427 for PASTE, 3380.5 for Scanorama, and 1852 for Seurat. Rank 1 is the highest rank.
Supplementary information
Rights and permissions
About this article
Cite this article
Zeira, R., Land, M., Strzalkowski, A. et al. Alignment and integration of spatial transcriptomics data. Nat Methods 19, 567–575 (2022). https://doi.org/10.1038/s41592-022-01459-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-022-01459-6
This article is cited by
-
Multi-slice spatial transcriptome domain analysis with SpaDo
Genome Biology (2024)
-
Streamlining spatial omics data analysis with Pysodb
Nature Protocols (2024)
-
Cellstitch: 3D cellular anisotropic image segmentation via optimal transport
BMC Bioinformatics (2023)
-
SPIRAL: integrating and aligning spatially resolved transcriptomics data across different experiments, conditions, and technologies
Genome Biology (2023)
-
Construction of a 3D whole organism spatial atlas by joint modelling of multiple slices with deep neural networks
Nature Machine Intelligence (2023)



