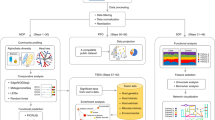
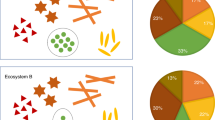
Abstract
Mako is a software tool that converts microbiome data and networks into a graph database and visualizes query results, thus allowing users without programming knowledge to carry out network-based queries. Mako is accompanied by a database compiled from 60 microbiome studies that is easily extended with the user’s own data. We illustrate mako’s strengths by enumerating association partners linked to propionate production and comparing frequencies of different network motifs across habitat types.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
The complete Neo4j database used in this paper has been used to generate a Neo4j dump file. An archived version of this file has been submitted to Zenodo17. Instructions for reconstructing this database are available in mako’s documentation. All BIOM files were downloaded from QIITA6 and a full overview of these studies can be found in Supplementary Table 1.
Code availability
The latest version of the mako software can be downloaded via https://github.com/ramellose/mako/. An archived version, including the script used to quantify motifs in the Neo4j database, has been submitted to Zenodo17. All mako source code and the included script are available under the Apache License 2.0. An extensive manual for running mako and Neo4j (via Docker) is available via https://ramellose.github.io/mako_docs/. Additionally, an accompanying compute capsule allows a demo of mako and a Neo4j database to be run without requiring any installations21.
Change history
23 March 2022
A Correction to this paper has been published: https://doi.org/10.1038/s41592-022-01462-x
References
Röttjers, L. & Faust, K. From hairballs to hypotheses–biological insights from microbial networks. FEMS Microbiol. Rev. 42, 761–780 (2018).
Jackson, M. A. et al. Detection of stable community structures within gut microbiota co-occurrence networks from different human populations. PeerJ 6, e4303 (2018).
Wang, H. et al. Combined use of network inference tools identifies ecologically meaningful bacterial associations in a paddy soil. Soil Biol. Biochem. 105, 227–235 (2017).
Poisot, T. et al. mangal–making ecological network analysis simple. Ecography 39, 384–390 (2016).
Szklarczyk, D. et al. String v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 (2019).
Gonzalez, A. et al. QIITA: rapid, web-enabled microbiome meta-analysis. Nat. Methods 15, 796–798 (2018).
Miller, J. J. Graph database applications and concepts with neo4j. In Proc. Southern Association for Information Systems Conference, Atlanta, GA, USA Vol. 2324 (ed. Fitzpatrick, L.) 141–147 (AIS, 2013).
Bansal, S. K. Towards a semantic extract-transform-load (ETL) framework for big data integration. In Proc. 2014 IEEE International Congress on Big Data (eds Chen, P. & Jain, H.) 522–529 (IEEE, 2014).
Noy, N. F. et al. Creating semantic web contents with protege-2000. IEEE Intell. Syst. 16, 60–71 (2001).
Kerr, B., Riley, M. A., Feldman, M. W. & Bohannan, B. J. Local dispersal promotes biodiversity in a real-life game of rock–paper–scissors. Nature 418, 171–174 (2002).
Ma, Z. S. & Ye, D. Trios-promising in silico biomarkers for differentiating the effect of disease on the human microbiome network. Sci. Rep. 7, 13259 (2017).
Thompson, L. R. et al. A communal catalogue reveals earth’s multiscale microbial diversity. Nature 551, 457–463 (2017).
Ma, B. et al. Earth microbial co-occurrence network reveals interconnection pattern across microbiomes. Microbiome 8, 82 (2020).
Faust, K. et al. Cross-biome comparison of microbial association networks. Front. Microbiol. 6, 1200 (2015).
Louis, P. & Flint, H. J. Formation of propionate and butyrate by the human colonic microbiota. Environ. Microbiol. 19, 29–41 (2017).
Tackmann, J., Rodrigues, J. F. M. & von Mering, C. Rapid inference of direct interactions in large-scale ecological networks from heterogeneous microbial sequencing data. Cell Syst. 9, 286–296 (2019).
Röttjers, L. & Faust, K. Fast and flexible analysis of linked microbiome data with mako. Zenodo https://doi.org/10.5281/zenodo.4946425 (2021).
Conway, J. R., Lex, A. & Gehlenborg, N. Upsetr: an R package for the visualization of intersecting sets and their properties. Bioinformatics 33, 2938–2940 (2017).
Sioutos, N. et al. Nci thesaurus: a semantic model integrating cancer-related clinical and molecular information. J. Biomed. Inform. 40, 30–43 (2007).
Summer, G. et al. cyneo4j: connecting neo4j and cytoscape. Bioinformatics 31, 3868–3869 (2015).
Röttjers, L. & Faust, K. Fast and flexible analysis of linked microbiome data with mako. Code Ocean https://doi.org/10.24433/CO.0482418.v1 (2021).
Acknowledgements
This project was supported by the KU Leuven under grant no. STG/16/006. K.F. has received funding from the European Research Council under the European Union’s Horizon 2020 research and innovation program under grant agreement no. 801747.
Author information
Authors and Affiliations
Contributions
L.R. developed the software, carried out the case studies and drafted the paper. K.F. contributed to the design of the software and the case studies, and provided substantial revisions of the paper. Both authors read and approved the final paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Methods thanks Jianguo Xia and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available. Lin Tang was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Table 1
Excel file containing a summary of all downloaded BIOM files and derived network size (in terms of node number and edge number) used for the study ‘Fast and flexible analysis of linked microbiome data with mako’.
Rights and permissions
About this article
Cite this article
Röttjers, L., Faust, K. Fast and flexible analysis of linked microbiome data with mako. Nat Methods 19, 51–54 (2022). https://doi.org/10.1038/s41592-021-01335-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-021-01335-9
This article is cited by
-
Karoline Faust
Nature Methods (2022)
-
Enhancing biomarkers with co-abundance
Nature Computational Science (2022)