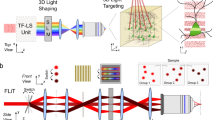
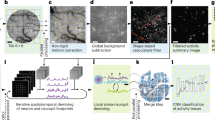
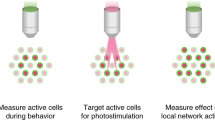
Abstract
Optical recording of neuronal activity in three-dimensional (3D) brain circuits at cellular and millisecond resolution in vivo is essential for probing information flow in the brain. While random-access multiphoton microscopy permits fast optical access to neuronal targets in three dimensions, the method is challenged by motion artifacts when recording from behaving animals. Therefore, we developed three-dimensional custom-access serial holography (3D-CASH). Built on a fast acousto-optic light modulator, 3D-CASH performs serial sampling at 40 kHz from neurons at freely selectable 3D locations. Motion artifacts are eliminated by targeting each neuron with a size-optimized pattern of excitation light covering the cell body and its anticipated displacement field. Spike rates inferred from GCaMP6f recordings in visual cortex of awake mice tracked the phase of a moving bar stimulus with higher spike correlation between intra compared to interlaminar neuron pairs. 3D-CASH offers access to the millisecond correlation structure of in vivo neuronal activity in 3D microcircuits.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Data used for the main figures are available on https://www.opendata.bio.ens.psl.eu/3D-CASH/. Further data are available from the authors upon request.
Code availability
Codes for hologram retrieval are available on https://github.com/walther007/3D-CASH. Other codes will be provided by the authors upon request.
Change history
25 January 2022
A Correction to this paper has been published: https://doi.org/10.1038/s41592-022-01401-w
References
Svoboda, K. & Yasuda, R. Principles of two-photon excitation microscopy and its applications to neuroscience. Neuron 50, 823–839 (2006).
Dombeck, D. A., Khabbaz, A. N., Collman, F., Adelman, T. L. & Tank, D. W. Imaging large-scale neural activity with cellular resolution in awake, mobile mice. Neuron 56, 43–57 (2007).
Göbel, W., Kampa, B. M. & Helmchen, F. Imaging cellular network dynamics in three dimensions using fast 3D laser scanning. Nat. Methods 4, 73–79 (2007).
Grewe, B. F., Voigt, F. F., van’t Hoff, M. & Helmchen, F. Fast two-layer two-photon imaging of neuronal cell populations using an electrically tunable lens. Biomed. Opt. Express 2, 2035–2046 (2011).
Kong, L. et al. Continuous volumetric imaging via an optical phase-locked ultrasound lens. Nat. Methods 12, 759–762 (2015).
Yang, W. et al. Simultaneous multi-plane imaging of neural circuits. Neuron 89, 269–284 (2016).
Botcherby, E. J. et al. Aberration-free three-dimensional multiphoton imaging of neuronal activity at kHz rates. Proc. Natl Acad. Sci. 109, 2919–2924 (2012).
Liu, R. et al. Aberration-free multi-plane imaging of neural activity from the mammalian brain using a fast-switching liquid crystal spatial light modulator. Biomed. Opt. Express 10, 5059–5080 (2019).
Quirin, S., Peterka, D. S. & Yuste, R. Instantaneous three-dimensional sensing using spatial light modulator illumination with extended depth of field imaging. Opt. Express 21, 16007–16021 (2013).
Ducros, M., Houssen, Y. G., Bradley, J., de Sars, V. & Charpak, S. Encoded multisite two-photon microscopy. Proc. Natl Acad. Sci. 110, 13138–13143 (2013).
Thériault, G., Cottet, M., Castonguay, A., McCarthy, N., & De Koninck, Y. Extended tw-photon microscopy in live samples with Bessel beams: steadier focus, faster volume scans, and simpler stereoscopic imaging. Front. Cell. Neurosci. 8, 1–11 (2014).
Song, A. et al. Volumetric two-photon imaging of neurons using stereoscopy (vTwINS). Nat. Methods 14, 420–426 (2017).
Cheng, A., Gonçalves, J. T., Golshani, P., Arisaka, K. & Portera-Cailliau, C. Simultaneous two-photon calcium imaging at different depths with spatiotemporal multiplexing. Nat. Methods 8, 139–142 (2011).
Picot, A. et al. Temperature rise under two-photon optogenetic brain stimulation. Cell Rep. 24, 1243–1253.e5 (2018).
Podgorski, K. & Ranganathan, G. Brain heating induced by near-infrared lasers during multiphoton microscopy. J. Neurophysiol. 116, 1012–1023 (2016).
Bullen, A., Patel, S. S. & Saggau, P. High-speed, random-access fluorescence microscopy: I. High-resolution optical recording with voltage-sensitive dyes and ion indicators. Biophys. J. 73, 477–491 (1997).
Salomé, R. et al. Ultrafast random-access scanning in two-photon microscopy using acousto-optic deflectors. J. Neurosci. Methods 154, 161–174 (2006).
Reddy, G. D. & Saggau, P. Fast three-dimensional laser scanning scheme using acousto-optic deflectors. J. Biomed. Opt. 10, 1–10 (2005).
Otsu, Y. et al. Optical monitoring of neuronal activity at high frame rate with a digital random-access multiphoton (RAMP) microscope. J. Neurosci. Methods 173, 259–270 (2008).
Reddy, G., Kelleher, K., Fink, R. & Saggau, P. Three-dimensional random access multiphoton microscopy for functional imaging of neuronal activity. Nat. Neurosci. 11, 713–720 (2008).
Katona, G. et al. Fast two-photon in vivo imaging with three-dimensional random-access scanning in large tissue volumes. Nat. Methods 9, 201–208 (2012).
Fernández-Alfonso, T. et al. Monitoring synaptic and neuronal activity in 3D with synthetic and genetic indicators using a compact acousto-optic lens two-photon microscope. J. Neurosci. Methods 222, 69–81 (2014).
Grewe, B. F., Langer, D., Kasper, H., Kampa, B. M. & Helmchen, F. High-speed in vivo calcium imaging reveals neuronal network activity with near-millisecond precision. Nat. Methods 7, 399–405 (2010).
Szalay, G. et al. Fast 3D imaging of spine, dendritic, and neuronal assemblies in behaving animals. Neuron 92, 723–738 (2016).
Griffiths, V. A. et al. Real-time 3D movement correction for two-photon imaging in behaving animals. Nat. Methods 17, 741–748 (2020).
Akemann, W. et al. Fast spatial beam shaping by acousto-optic diffraction for 3D non-linear microscopy. Opt. Express 23, 28191–28205 (2015).
Kaplan, A., Friedman, N. & Davidson, N. Acousto-optic lens with very fast focus scanning. Opt. Lett. 26, 1078–1080 (2001).
Botcherby, E. J., Juškaitis, R., Booth, M. J. & Wilson, T. An optical technique for remote focusing in microscopy. Opt. Commun. 281, 880–887 (2008).
Wyrowski, F. & Bryngdahl, O. Iterative Fourier-transform algorithm applied to computer holography. J. Opt. Soc. Am. A 5, 1058–1065 (1988).
Donnert, G., Eggeling, C. & Hell, S. W. Major signal increase in fluorescence microscopy through dark-state relaxation. Nat. Methods 4, 81–86 (2007).
Dipoppa, M. et al. Vision and locomotion shape the interactions between neuron types in mouse visual cortex. Neuron 98, 602–615.e8 (2018).
Kerr, J. N. D. et al. Spatial organization of neuronal population responses in layer 2/3 of rat barrel cortex. J. Neurosci. 27, 13316–13328 (2007).
Smith, S. L. & Häusser, M. Parallel processing of visual space by neighboring neurons in mouse visual cortex. Nat. Neurosci. 13, 1144–1149 (2010).
Saleem, A. B., Ayaz, A., Jeffery, K. J., Harris, K. D. & Carandini, M. Integration of visual motion and locomotion in mouse visual cortex. Nat. Neurosci. 16, 1864–1869 (2013).
Deneux, T. et al. Accurate spike estimation from noisy calcium signals for ultrafast three-dimensional imaging of large neuronal populations in vivo. Nat. Commun. 7, 12190 (2016).
Chen, T.-W. et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300 (2013).
Niell, C. M. & Stryker, M. P. Modulation of visual responses by behavioral state in mouse visual cortex. Neuron 65, 472–479 (2010).
Tubiana, J., Wolf, S., Panier, T. & Debregeas, G. Blind deconvolution for spike inference from fluorescence recordings. J. Neurosci. Methods 342, 108763 (2020).
Nadella, K. M. N. S. et al. Random-access scanning microscopy for 3D imaging in awake behaving animals. Nat. Methods 13, 1001–1004 (2016).
Kirkby, P. A., Srinivas Nadella, K. M. N. & Silver, R. A. A compact acousto-optic lens for 2D and 3D femtosecond based 2-photon microscopy. Opt. Express 18, 13720–13744 (2010).
Weisenburger, S. et al. Volumetric Ca2+ imaging in the mouse brain using hybrid multiplexed sculpted light microscopy. Cell 177, 1050–1066.e14 (2019).
Zhang, T. et al. Kilohertz two-photon brain imaging in awake mice. Nat. Methods 16, 1119–1122 (2019).
Kazemipour, A. et al. Kilohertz frame-rate two-photon tomography. Nat. Methods 16, 778–786 (2019).
Wu, J. et al. Kilohertz two-photon fluorescence microscopy imaging of neural activity in vivo. Nat. Methods 17, 287–290 (2020).
de Vries, S. E. J. et al. A large-scale standardized physiological survey reveals functional organization of the mouse visual cortex. Nat. Neurosci. 23, 138–151 (2020).
Ohki, K., Chung, S., Ch’ng, Y. H., Kara, P. & Reid, R. C. Functional imaging with cellular resolution reveals precise micro-architecture in visual cortex. Nature 433, 597–603 (2005).
Skottun, B. C. et al. Classifying simple and complex cells on the basis of response modulation. Vis. Res. 31, 1078–1086 (1991).
Yunzab, M., Cloherty, S. L. & Ibbotson, M. R. Comparison of contrast-dependent phase sensitivity in primary visual cortex of mouse, cat and macaque. NeuroReport 30, 960–965 (2019).
Niell, C. M. & Stryker, M. P. Highly selective receptive fields in mouse visual cortex. J. Neurosci. 28, 7520–7536 (2008).
Chance, F. S., Nelson, S. B. & Abbott, L. F. Complex cells as cortically amplified simple cells. Nat. Neurosci. 2, 277–282 (1999).
Bonin, V., Histed, M. H., Yurgenson, S. & Reid, R. C. Local diversity and fine-scale organization of receptive fields in mouse visual cortex. J. Neurosci. 31, 18506–18521 (2011).
Xu, J. & Stroud, R. Acousto-Optic Devices (Wiley, 1992).
Kremer, Y. et al. A spatio-temporally compensated acousto-optic scanner for two-photon microscopy providing large field of view. Opt. Express 16, 10066–10076 (2008).
Beaurepaire, E. & Mertz, J. Epifluorescence collection in two-photon microscopy. Appl. Opt. 41, 5376–5382 (2002).
Oheim, M., Beaurepaire, E., Chaigneau, E., Mertz, J. & Charpak, S. Two-photon microscopy in brain tissue: parameters influencing the imaging depth. J. Neurosci. Methods 111, 29–37 (2001).
Striker, G., Subramaniam, V., Seidel, C. A. M. & Volkmer, A. Photochromicity and fluorescence lifetimes of green fluorescent protein. J. Phys. Chem. B 103, 8612–8617 (1999).
Antonini, A., Liberale, C. & Fellin, T. Fluorescent layers for characterization of sectioning microscopy with coverslip-uncorrected and water immersion objectives. Opt. Express 22, 14293–14304 (2014).
Pnevmatikakis, E. A. & Giovannucci, A. NoRMCorre: an online algorithm for piecewise rigid motion correction of calcium imaging data. J. Neurosci. Methods 291, 83–94 (2017).
Erdélyi, F. et al. Preferential origin and layer destination of GAD65-GFP cortical interneurons. Cereb. Cortex 14, 1122–1133 (2004).
Villette, V., Levesque, M., Miled, A., Gosselin, B. & Topolnik, L. Simple platform for chronic imaging of hippocampal activity during spontaneous behaviour in an awake mouse. Sci. Rep. 7, 43388 (2017).
Reimer, J. et al. Pupil fluctuations track rapid changes in adrenergic and cholinergic activity in cortex. Nat. Commun. 7, 13289 (2016).
Acknowledgements
We wish to acknowledge expert technical help by G. Paresys for electronic designs, Y. Cabirou for mechanical engineering, E. M. Afif for advice on FPGA programming, F. Le Fouiller for explanations of AO device design, the IBENS transgenesis platform and C. Maihes-Hamon for help with virus injections, C. Dussaux (Institut Pasteur, Paris) for spin-coated fluorescent micro-slides, P. Rostaing for ultramicrotome sectioning of fluorescent nanoslides, C. Auger for help with pharmacology and the IBENS animal facility for animal care. We also want to thank N. Rochefort, F. Chavane, S. Gigan and D. Ferster for discussions. Funding was provided by ANR (grant nos ALPINS ANR-15-CE19-0011-01, ANR-15-CE19-0011-02 and EXPECT ANR-17-CE37-0022-01), Région Île de France (DIM Cerveau & Pensée, ALPINS), NIH BRAIN Initiative (grant no. 1U01NS103464), Fondation pour l’Audition (grant no. FPA RD-2018-6), Institut de Convergence Q-life (grant no. Q-life ANR-17-CONV-0005) and the program Investissements d’Avenir launched by the French government and implemented by the ANR, with the references: ANR-10-LABX-54 (Memolife), ANR-10-IDEX-0001-02 (Université PSL) and ANR-10-INSB-04- 01 (France-BioImaging Infrastructure). S.W. was supported by a postdoctoral fellowship SPF201809007064 from the Fondation pour la Recherche Médicale France.
Author information
Authors and Affiliations
Contributions
L.B., S.D. and J.-F.L. initiated research and obtained funding. W.A. built the 3DScope. W.A. and B.M. coded the software. B.M. developed the user interface. J.F. and W.A. designed the detection unit. A.T. set up the transmission microscope. J.F. designed the tower head post. V.V. set up the treadmill. W.A. set up the visual stimulator and isoflurane anesthesia. V.V. and W.A. performed animal surgery. V.V. performed virus injections. W.A. performed experiments. S.D. and V.V. performed cell-attached recordings. W.A. and S.W. analyzed data and created figures. L.B., S.D., J.-F.L., C.V., V.V., S.W. and W.A. discussed research. W.A., L.B., S.D. and J.-F.L. wrote the manuscript with contributions from all authors.
Corresponding authors
Ethics declarations
Competing interests
B.M. and S.D. are cofounders of the company Karthala System and hold shares in this company. J.-F.L., L.B. and S.D. are inventors in accordance with U.S. patents 10423017 and 10191268 jointly owned by the Institut National de la Santé et de la Recherche Médicale (INSERM), the Centre National de la Recherche Scientifique (CNRS) and the École Normale Supérieure de Paris (ENS). All other authors declare no competing interests.
Additional information
Peer review information Nature Methods thanks Meng Cui, Takafumi Inoue and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Nina Vogt was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Optical hardware (3DScope).
(a) Schematic representation of the active components in the signal and idler paths of the microscope (PMT: photomultiplier tube detector; AOD: acousto-optic deflector; AOM: acousto-optic modulator; OPA: optical parametric amplifier; CPA: chirped pulse amplifier). (b-d) 3D-rendered views of the microscope. (b) Top view without table and safety walls. (c) Microscope head with XYZ-movable objective (modified Movable Objective Microscope, MOM, Sutter Instruments) with added 2″ periscope and custom-designed relay optics and detection unit. (d) Complete view with transparent safety walls for the purpose of visualization.
Extended Data Fig. 2 Simultaneous CASH and image-scan recording.
(a) Experimental design: A single GCaMP6f-expressing cell is targeted first with a cell-centered image scan (39×39 pixels, 38 ms scan time) followed by a two-shot CASH acquisition (50 μs) resulting in 26 Hz loop rate. (b) Example recording (130 s) of spontaneous activity showing (top to bottom) the motion of the animal, absolute fluorescence signal obtained from 5×5 CASH (green), imaging (black; pixel mean after image stack alignment), the brightest pixel of the aligned image stack (blue), the 2×5 CASH signal (green) and the imaging-derived neuropil signal (black; pixel mean across a concentric region around the cell body). (c) Top: Overlay of ΔF/F fluorescence traces after neuropil (NP) correction and normalization minimizing the global mean squared difference. The alignment yields 0.98 coefficient of correlation (cc). Middle: amplitudes of cell motion in x and y direction from alignment of image frames. Bottom: fluorescence images of the targeted cell at different instants as marked above (black lines). (d) Overlay of the single pixel trace before (blue) and after (black) x,y-alignment of the time lapse image sequence. (e) Overlay of a CASH (green) and imaging (black; same cell as above), but with 810 nm excitation for Ca2+-independent fluorescence (bottom), with x,y cell motion (middle) and running speed of the animal (top). (f) Top: overlay of single-trial CASH (green) and imaging (black) recordings (different cell) during presentation of a visual noise stimulus (gray bars) yielding 0.94 correlation. Middle: x,y cell motions. Bottom: Snapshot fluorescence images of the cell at instants indicated by bars above (black). (g) 2D representation of x,y cell motion during recordings in (b; left) and (f; right) with moving speed of the animal in false color. (h) Coefficient of correlation of GCaMP6f traces of simultaneous CASH and imaging recordings, analogous to (b) and (f), for all recorded cells (violet bars; median) and of the same traces after removal of photon noise by MLspike deconvolution fit (green bars; median). The number of observed cells are indicated above. Error bars represent SD.
Extended Data Fig. 3 Event detection accuracy evaluated on simulated data.
a) Workflow schematic: We generated GCaMP6f-like neural calcium activity (racq = 250 Hz, 600 spikes, mean firing rate 0.2 Hz, 3000 s) contaminated with a neuropil signal. After neuropil correction, we deconvoluted the data and from inferred spikes evaluated the error rate (ER), the sensitivity and the precision of inference. (b) Error rates for artificial data (SNR 2, 3 and 4) according to neuropil correction (noise- or correlation regression) and inference algorithm (MLspike and BSD). (c) Error rate as function of acquisition rate, racq, for data with SNR between two and five obtained with the BSD algorithm for two window sizes ΔTwin (12 and 20 ms). A spike is rated as detected, if the spike falls within this time window centered around the time of the true spike.
Extended Data Fig. 4 Simultaneous CASH and cell-attached patch recording.
(a) Image of patch electrode-targeted neuron expressing GCaMP6f (green) and membrane-attached electrode filled with Alexa 594 (red). (b) Five minute example recording from the neuron shown in (a) with GCaMP6f signal (black), MLspike-retrieved GCaMP6f signal (green), electrode current at −65 mV holding potential (blue), position of spikes inferred from the current trace (red), spikes obtained from MLspike deconvolution (black) and MLspike-estimated spike probability (magenta). (c) Spike-sorted GCaMP6f transients in black (first row) and electrode-measured spikes in blue (second row) for single (1 AP; 21 events), double (2 AP; 14 events) and triple (3 AP; 19 events) action potential events with the first spike at time zero and drawn from 23 minutes total recording time. Detection was subject to the condition that no further event is found during 0.6 s before the first spike and 0.7 s after the last spike. The incidence window was 30 ms. (d) MLSpike-estimated spike probability (black crosses) for events shown in (c) in the −0.3 to 1.5 s from the electrode-detected spike event at time zero, together with the normalized cumulative probability (red) and probability density calculated with a gaussian kernel (20 ms; blue). Dotted curve (red) represents the cumulative ensemble probability at ± SD confidence (dotted red band) bootstrapped from segments of the recordings (104 repetitions) where no spikes were detected. (e) Diagnostic of optical spike detection for 1–3 AP events as function of a probability threshold with a 0.2 s detection window following an electrode-detected spike. The true positive (hit) and false negative (misses) rate was calculated from the MLspike-detected events reaching a probability beyond threshold anywhere within the detection window. (f) Spike-triggered average of the GCaMP6f transient for 1 AP (8 events), 2 AP (6 events) and 3 AP (19 events) which surpassed a 0.4 probability threshold (red) with ± 1 SD (red band), biexponential fit to the mean (green; 0.33 ± 0.4 decay time constant) and the ± 1 SD bootstrap distribution of GCaMP6f transients in absence of spike events (blue). (g) Mean GCaMP6f response amplitude for 1–3 AP events obtained from the thresholded spike response in (f), together with polynomial fit (red). (h) Left: Total number of electrode-detected spike events (true events) and MLspike-detected events (positive events). Right: Mean firing rate across the whole data set evaluated from true or from positive events, respectively. (i) Left: True positives (TP), false negatives (FN) and false positives (FP) counts across the whole data set when applying a 0.2 s detection window. Right: MLSpike inference sensitivity, precision and error rate evaluated from the counts in (h, left) and (I left).
Extended Data Fig. 5 Example cell with orientation-tuned response.
GCaMP6f orientation-sensitive visual responses (cell # 24 in Fig. 5d–f). (a) GCaMP6f fluorescence signal in response to the repeated presentation (20 repeats) of the same stimulus series (gray bars) with orientations indicated on the top of the figure. The moving speed of the animal is shown for every trial as a false-color bar together with the GCaMP6f trace (black). (b) Global orientation sensitivity index (gOSI) derived from single trials (blue dots) and from the trial average (red dot). (c) gOSI amplitude for all trials.
Extended Data Fig. 6 Single trial visual responses in a cell population.
(a) 3D coordinates of cells in the dataset (same as in Fig. 5b–h). (b) GCaMP6f fluorescence images of all cells in the dataset. (c) False-color representation of GCaMP6f fluorescence responses to visual contrast grating stimuli (vertical dotted lines). Shown are the responses in trials no 1, 3, 6 and 19 of a total of 20 trials (trial average shown in Fig. 5b) together with the locomotion velocity as horizontal color bar. The orientation of stimulus gratings, same for all trials, are shown on the very top.
Extended Data Fig. 7 GCaMP6f responses of V1 cells in the awake versus the anesthetized state.
(a) Fluorescence images of 36 cells (L2/3; two experiments; one animal) that passed the test for significant tuning response in both states (p≤0.03, bootstrap test). Values below indicate the z position below pia. (b) Single-trial fluorescence responses of cell #31 to the preferred orientation (150°) in the awake state (left) and in the anesthetized state under 0.25 % isoflurane (right). Traces in the awake state are color-coded according the maximum speed of locomotion of the animal. (c) Single-trial fluorescence responses of 24 cells by choosing for each cell the stimulus trial of maximum integral response in the awake (left) and anesthetized (right) state, using the same color code as in (b). (d) Overlaid histograms of response amplitude for the best stimulus across all trials and all cells in the awake (red) and anesthetized (gray) state. Responses with amplitude above 0.15 have low counts and are omitted from the figure. The zero response bar was cropped at the top. (e) Measured cell-averaged response probability in single trials (mean; 10 repetitions; SD error). Trial responses to the best stimulus, as judged from the average response, were considered as positive when their amplitude was at least three times larger than baseline noise, otherwise as failures. Number of animals given in brackets. (f) Trail-averaged integral response amplitude for the best stimulus for all cells when recorded in the awake (red) and anesthetized (black) state. Connecting lines indicate data belonging to the same cell with red lines for cells featuring larger response in the awake state. (g) Standard Deviation (SD) of trial responses for the best stimulus normalized to the mean of the response for cells when recorded in the awake (red) and (black) anesthetized state. Lines connect data belonging to the same cell with red lines for cells showing increased variance under isoflurane.
Extended Data Fig. 8 Spontaneous Ca2+ activity in the awake versus anesthetized state.
(a) 3D coordinates of cells in the dataset (47 cells). (b) GCaMP6f Fluorescence images of all cells. (c) Fluorescence traces in false colors, according to the color bar on the right side of (e) recorded in the awake state. Color bar on the top indicates locomotion velocity of the animal. (d) Spikes found in (c) by MLspike. (e) Fluorescence traces from the same cells as in (c), but with the mouse under 0.25 % isoflurane anesthesia. (f) Spikes found in (e) by MLspike. (g) Histogram of pairwise spike correlations in (d) between all cells (black), between layer 2/3 cells (red) and between layer 5 cells (blue). Spikes were counted as synchronous if they fall within a 10 ms time window. (h) Same as in (g), but evaluated from the dataset in (f). (i) Mean spike rates calculated from the spike maps in (d) and (f). Paired t-test, p = 2·10−4 (***; 47 cells), 2·10−3 (**; 36 cells), 1·10−2 (*; 10 cells). Pairs of same cells connected by lines (gray).
Extended Data Fig. 9 Example cell with phase-sensitive response.
(a) GCaMP6f response of a layer 2/3 example cell to a stimulus series of contrast gratings with grating orientations between 0° and 360° randomized between trials and moving at a speed of 2 cycles per second (cps) under isoflurane (0.25–0.5 %) anesthesia. Sampling with 363 Hz. Top: Trial #3. Bottom: Trial # 81 (Gaussian-filtered, sigma 2.5 ms). (b) Responses to 180° stimulus orientation of trial #3 (left) and # 81 (right) with MLspike-infered spikes indicated as black vertical marks (bottom) and convolution of inferred spikes with the GCaMP6f response template (green curve). (c) False-color representation of the GCaMP6f responses to the 180° (left) and 0° counterpropagating stimulus (right) for 88 consecutive trials together with MLspike-inferred spikes (blue vertical lines). (d) Stacked peristimulus time histogram (PSTH) obtained from the data in (c) with 80 ms binning width. The R1/R0 ratio of the data is 0.61.
Extended Data Fig. 10 Orientation and phase sensitive responses.
(a) Orientation sensitivity index (OSI) for four example neurons with orientation preferences (Θpref) 178°, 78°, 171° and 12° (top to bottom). Experimental (black) with biphasic fit (red). Error bars indicate SEM. (b) Stacked peristimulus time histograms (PSTH) obtained for the neurons in (a) from 80 (# 1, #2, #4) or 50 (# 3) trials with 80 ms binning width resulting in R1/R0 ratios ranging from 1.2 to 0.04 (top to bottom). (c) Harmonic spike Rate R1 as function of the mean rate R0 in response to the preferred stimulus orientation and preferred direction for 155 neurons in layer 2/3. Responses classified according to R1/R0 larger (red) or lower (blue) than the R1/R0 ensemble mean. Linear fits to the data given as dashed lines. (d) Same as (a) for the counterpropagating direction of the preferred stimulus.
Supplementary information
Supplementary Information
Supplementary Figs. 1–12 and Discussion.
Rights and permissions
About this article
Cite this article
Akemann, W., Wolf, S., Villette, V. et al. Fast optical recording of neuronal activity by three-dimensional custom-access serial holography. Nat Methods 19, 100–110 (2022). https://doi.org/10.1038/s41592-021-01329-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-021-01329-7
This article is cited by
-
Two-photon calcium imaging of neuronal activity
Nature Reviews Methods Primers (2022)