Abstract
Two-photon microscopy has enabled high-resolution imaging of neuroactivity at depth within scattering brain tissue. However, its various realizations have not overcome the tradeoffs between speed and spatiotemporal sampling that would be necessary to enable mesoscale volumetric recording of neuroactivity at cellular resolution and speed compatible with resolving calcium transients. Here, we introduce light beads microscopy (LBM), a scalable and spatiotemporally optimal acquisition approach limited only by fluorescence lifetime, where a set of axially separated and temporally distinct foci record the entire axial imaging range near-simultaneously, enabling volumetric recording at 1.41 × 108 voxels per second. Using LBM, we demonstrate mesoscopic and volumetric imaging at multiple scales in the mouse cortex, including cellular-resolution recordings within ~3 × 5 × 0.5 mm volumes containing >200,000 neurons at ~5 Hz and recordings of populations of ~1 million neurons within ~5.4 × 6 × 0.5 mm volumes at ~2 Hz, as well as higher speed (9.6 Hz) subcellular-resolution volumetric recordings. LBM provides an opportunity for discovering the neurocomputations underlying cortex-wide encoding and processing of information in the mammalian brain.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The raw image data presented in this work is currently too large for sharing via typical public repositories. It is available from the corresponding author upon reasonable request. Source data are provided with this paper.
Code availability
Stimulus delivery and treadmill control was implemented with a combination of MATLAB, Python, and Arduino scripts. Neuronal segmentation and non-rigid motion correction were based on the CaImAn53,54 and NoRMCorre52 software packages, respectively, and implemented using MATLAB. All custom code, including pipelines based on CaImAn and NoRMCorre, is publicly available on the Vaziri lab GitHub repository (https://github.com/vazirilab).
Change history
08 November 2021
A Correction to this paper has been published: https://doi.org/10.1038/s41592-021-01337-7
References
Denk, W., Strickler, J. H. & Webb, W. W. Two-photon laser scanning fluorescence microscopy. Science 248, 73–76 (1990).
So, P. T. C., Dong, C. Y., Masters, B. R. & Berland, K. M. Two photon excitation fluorescence microscopy. Annu. Rev. Biomed. Eng. 2, 399–429 (2000).
Helmchen, F. & Denk, W. Deep tissue two-photon microscopy. Nat. Methods 2, 932–940 (2005).
Miyawaki, A. et al. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature 388, 882–887 (1997).
Nakai, J., Ohkura, M. & Imoto, K. A high signal-to-noise Ca2+ probe composed of a single green fluorescent protein. Nat. Biotechnol. 19, 137–141 (2001).
Chen, T. et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300 (2013).
Harris, J. A. et al. Hierarchical organization of cortical and thalamic connectivity. Nature 575, 195–202 (2019).
Kuchibhotla, K. V. et al. Parallel processing by cortical inhibition enables context-dependent behavior. Nat. Neurosci. 20, 62–71 (2017).
Stringer, C., Pachitariu, M., Steinmetz, N., Carandini, M. & Harris, K. D. High-dimensional geometry of population responses in visual cortex. Nature 571, 361–365 (2019).
Rumyantsev, O. I. et al. Fundamental bounds on the fidelity of sensory cortical coding. Nature 580, 100–105 (2020). NC.
Musall, S., Kaufman, M. T., Juavinett, A. L., Gluf, S. & Churchland, A. K. Single-trial neural dynamics are dominated by richly varied movements. Nat. Neurosci. 22, 1677–1686 (2019). NC.
Makino, H. et al. Transformation of cortex-wide emergent properties during motor learning. Neuron 94, 880–890 (2017).
Mao, T. et al. Long-range neuronal circuits underlying the interaction between sensory and motor cortex. Neuron 71, 111–123 (2011).
Pinto, L. et al. Task-dependent changes in the large-scale dynamics and necessity of cortical regions. Neuron 104, 810–8214 (2019).
Lin, Q. et al. Cerebellar neurodynamics predict decision timing and outcome on the Single-Trial level. Cell 180, 1–16 (2020).
Bartolo, R., Saunders, R. C., Mitz, A. R. & Averbeck, B. B. Information-limiting correlations in large neural populations. J. Neurosci. 40, 1668–1678 (2020).
Tsai, P. S. et al. Ultra-large field-of-view two-photon microscopy. Opt. Express 23, 13833–13847 (2015).
Sofroniew, N. J., Flickinger, D., King, J. & Svoboda, K. A large field of view two-photon mesoscope with subcellular resolution for in vivo imaging. eLife 5, 14472 (2016).
Stirman, J. N., Smith, I. T., Kudenov, M. W. & Smith, S. L. Wide field- of-view, multi-region, two-photon imaging of neuronal activity in the mammalian brain. Nat. Biotechnol. 34, 857–862 (2016).
Ji, N., Freeman, J. & Smith, S. L. Technologies for imaging neural activity in large volumes. Nat. Neurosci. 19, 1154–1164 (2016).
Weisenburger, S. & Vaziri, A. A guide to emerging technologies for large-scale and whole-brain optical imaging of neuronal activity. Annu. Rev. Neurosci. 41, 431–452 (2018).
Yang, W. & Yuste, R. In vivo imaging of neural activity. Nat. Methods 14, 349–359 (2017).
Yu, C., Stirman, J. N., Riichiro Hira, Y. Y., & Smith, S. L. Diesel2p mesoscope with dual independent scan engines for flexible capture of dynamics in distributed neural circuitry. Preprint at bioRxiv https://doi.org/10.1101/2020.09.20.305508 (2020).
Prevedel, R. et al. Fast volumetric calcium imaging across multiple cortical layers using sculpted light. Nat. Methods 13, 1021–1028 (2016).
Weisenburger, S. et al. Volumetric Ca2+ imaging in the mouse brain using hybrid multiplexed sculpted light (HyMS) microscopy. Cell 177, 1–17 (2019).
Kazemipour, A. et al. Kilohertz frame-rate two-photon tomography. Nat. Methods 16, 778–786 (2019).
Botcherby, E. J., Juškaitis, R. & Wilson, T. Scanning two photon fluorescence microscopy with extended depth of field. Opt. Commun. 268, 253–260 (2006).
Song, A. et al. Volumetric two-photon imaging of neurons using stereoscopy (vTwINS). Nat. Methods 14, 420–426 (2017).
Lu, R. et al. Rapid mesoscale volumetric imaging of neural activity with synaptic resolution. Nat. Methods 17, 291–294 (2020).
Zhang, T. et al. Kilohertz two-photon brain imaging in awake mice. Nat. Methods 16, 1119–1122 (2019).
Tsai, Y.-H. et al. Two-photon microscopy at >500 volumes/second. Preprint at bioRxiv https://doi.org/10.1101/2020.10.21.349712 (2020).
Amir, W. et al. Simultaneous imaging of multiple focal planes using a two-photon scanning microscope. Opt. Lett. 32, 1731–1733 (2007).
Cheng, A., Gonçalves, J., Golshani, P., Arisaka, K. & Portera-Cailliau, C. Simultaneous two-photon calcium imaging at different depths with spatiotemporal multiplexing. Nat. Methods 8, 139–142 (2011).
Tsyboulski, D. et al. Remote focusing system for simultaneous dual-plane mesoscopic multiphoton imaging. Preprint at bioRxiv https://doi.org/10.1101/503052 (2018).
Wu, J. et al. Ultrafast laser-scanning time-stretch imaging at visible wavelengths. Light.: Sci. Appl. 6, e16196 (2017).
Beaulieu, D. R., Davison, I. G., Kılıç, K., Bifano, T. G. & Mertz, J. Simultaneous multiplane imaging with reverberation two-photon microscopy. Nat. Methods 17, 283–286 (2020).
Wu, J. et al. Kilohertz two-photon fluorescence microscopy imaging of neural activity in vivo. Nat. Methods 17, 287–290 (2020).
Daigle, T. L. et al. A suite of transgenic driver and reporter mouse lines with enhanced brain-cell-type targeting and functionality. Cell 174, 465–480 (2018).
Horton, N. et al. In vivo three-photon microscopy of subcortical structures within an intact mouse brain. Nat. Photon 7, 205–209 (2013).
Cooper, B. G., Manka, T. F. & Mizumori, S. J. Y. Finding your way in the dark: the retrosplenial cortex contributes to spatial memory and navigation without visual cues. Behav. Neurosci. 115, 1012–1028 (2001).
Cohen, M. R. & Kohn, A. Measuring and interpreting neuronal correlations. Nat. Neurosci. 14, 811–819 (2011).
Stringer, C. et al. Spontaneous behaviors drive multidimensional, brainwide activity. Science 364, 255 (2019).
Dana, H. et al. High-performance calcium sensors for imaging activity in neuronal populations and microcompartments. Nat. Methods 16, 649–657 (2019).
Rigotti, M. et al. The importance of mixed selectivity in complex cognitive tasks. Nature 497, 585–590 (2013).
Abbott, L. F. & Dayan, P. The effect of correlated variability on the accuracy of a population code. Neural Comput. 11, 91–101 (1999). NC.
Steinmetz, N. A., Zatka-Haas, P., Carandini, M. & Harris, K. D. Distributed coding of choice, action and engagement across the mouse brain. Nature 576, 266–273 (2019).
Runyan, C., Piasini, E., Panzeri, S. & Harvey, C. D. Distinct timescales of population coding across cortex. Nature 548, 92–96 (2017).
Li, N., Daie, K., Svoboda, K. & Druckmann, S. Robust neuronal dynamics in premotor cortex during motor planning. Nature 532, 459–464 (2016).
Howe, M. W. & Dombeck, D. A. Rapid signalling in distinct dopaminergic axons during locomotion and reward. Nature 535, 505–510 (2016).
Rajasethupathy, P. et al. Projections from neocortex mediate top-down control of memory retrieval. Nature 526, 653–659 (2015).
Lau, C. et al. Exploration and visualization of gene expression with neuroanatomy in the adult mouse brain. BMC Bioinf. 9, 153–163 (2008).
Zariwala, H. A. et al. A Cre-dependent GCaMP3 reporter mouse for neuronal imaging in vivo. J. Neuroscience 32, 3131–3141 (2012).
Pnevmatikakis, E. A. & Giovannucci, A. NoRMCorre: an online algorithm for piecewise rigid motion correction of calcium imaging data. J. Neurosci. Meth. 291, 83–94 (2016).
Pnevmatikakis, E. A. et al. Simultaneous denoising, deconvolution, and demixing of calcium imaging data. Neuron 89, 285–299 (2016).
Giovannucci, A. et al. CaImAn: an open source tool for scalable calcium imaging data analysis. eLife 8, 38173 (2019).
Kandel, E. R., Schwartz, J. H. & Jessell, T. M. Principles of Neural Science. (McGraw-Hill, Health Professions Division, 2000).
Mathis, A. et al. DeepLabCut: markerless pose estimation of user-defined body parts with deep learning. Nat. Neurosci. 21, 1281–1289 (2018).
Nath, T. et al. Using DeepLabCut for 3D markerless pose estimation across species and behaviors. Nat. Protoc. 14, 2152–2176 (2019).
Stujenske, J. M., Spellman, T. & Gordon, J. A. Modeling the spatiotemporal dynamics of light and heat propagation for in vivo optogenetics. Cell Rep. 12, 525–534 (2015).
Podgorski, K. & Ranganathan, G. Brain heating induced by near-infrared lasers during multiphoton microscopy. J. Neurophysiol. 116, 1012–1023 (2016).
Acknowledgements
We thank P. Strogies and J. M. Petrillo (Precision Instrumentation Technology, Rockefeller University) for manufacturing mechanical components and K. Cialowicz (Bio-Imaging Resource Center, Rockefeller University) for performing confocal imaging of immunolabelled samples. We thank S. Weisenburger (LUMICKS) for helpful discussions related to microscope development and synchronization, and T. Nöbauer (Rockefeller University) for discussions regarding data management and processing. We thank K. Podgorski (Howard Hughes Medical Institute) for sharing previously used simulation software58 for laser-induced heating. Research reported in this publication was supported by the National Institute of Neurological Disorders and Stroke of the National Institutes of Health under award numbers 5U01NS103488, 1RF1NS113251, and 1RF1NS110501 (A. V.) and the Kavli Foundation (A.V., J. M., J. D.). This research was supported in part by a Bristol-Myers Squibb Postdoctoral Fellowship (J. D.).
Author information
Authors and Affiliations
Contributions
J. D. contributed to the project conceptualization, designed and built the imaging and data acquisition system, performed experiments, programmed experimental control and analysis software, analyzed data, and wrote the manuscript. J. M. performed data processing and modeling and contributed to writing the manuscript. F. T. contributed to microscope construction and characterization and aided in developing the experimental control design. H. K., F. M. T., and B. C. performed virus injections and cranial window surgeries. K. B. performed immunohistochemistry experiments. A. V. conceived and led the project, designed the imaging system, the data acquisition approach, and all in vivo mouse experiments, guided data analysis, and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
A. V. and J. D. described Light Beads Microscopy in patent application PCT/US2021/015957.
Additional information
Peer review information Nature Methods thanks Rosa Cossart and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
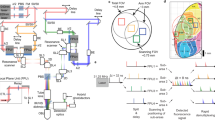
Extended Data Fig. 1 Full schematic of MAxiMuM.
a, Primary cavity schematic, where ‘Ms’ denote mirrors, ‘Ls’ denote lenses, and ‘HWP’ denotes a half-wave plate. Insets show a schematic of M1 in the Y-Z plane illustrating that the beam entering the cavity passes over top of M1, while subsequent beams encounter M1 and a camera image of the first ~7 beams exiting the cavity just after M1. b, A second cavity with shorter focal length mirrors creates a copy of the 15 pulses from Cavity A and shifts them in time and axial location to achieve the full 30 beams and 465 μm axial range of the MAxiMuM system. c, Temporal schematic of pulses from cavity A and B. Due to the shorter delay of cavity B relative to A, the pulse trains are interleaved. The pulse energies for each beam decrease exponentially due to the partially transmissive mirror (M1) in cavity A. Exponential decrease is matched to the expected scattering length (ls) for brain tissue (~200 μm). Power levels for cavity B pulses are lower than those from A since cavity B pulses are sent to more superficial layers in the brain; relative power can be controlled by the HWP in a. d, Sub-volume schematic. Cavities A and B form two sub-volumes, with the planes from cavity A below those from B such that together they continuously sample the entire axial range.
Extended Data Fig. 2 CAD drawings of the MAxiMuM system.
Scale bars: 100 mm.
Extended Data Fig. 3 Full microscope schematic.
a, Setup schematic for mesoscope system starting from the fiber chirped-pulse amplifier (FCPA), through the optical parametric chirped-pulse amplifier (OPCPA), electro-optic modulator (EOM), dispersion compensation path, MAxiMuM, and into the microscope. ‘Ls’ denote lenses, ‘Rs’ denote relay lens pairs, ‘PMT’ denotes photo-multiplier tube, ‘ADC’ denotes analog to digital converter, and ‘PLL’ denotes phase-locked loop. b, Schematic showing channel allocation for demultiplexing of signal from three adjacent light beads on the FPGA. Data points are the measured impulse response for fluorescence from GCaMP6f measured with our PMT and associated electronics, captured with 1614 MHz (0.62 ns) resolution. Shaded regions denote the integration boundaries for each de-multiplexed channel. c, Measurement of crosstalk of channels 1—15 into channels 16—30 (that is Cavity A → Cavity B). Black horizontal line shows mean value at ~7%.
Extended Data Fig. 4 Post-objective calibration of light bead columns generated by MAxiMuM.
a–d, Axial position (a), estimated pulse energy out of the objective (b) and in the sample (c), and transverse position of the light beads from MAxiMuM (d), calibrated by translating a pollen grain through the focus of the microscope. Estimated pulse energies assume 450 mW average power and a scattering length of 200 μm. e, Pulse duration measurements of each beam from MAxiMuM, post-objective. f-j, characterization of light bead point-spread functions. f, example images of PSFs for light beads 5, 15, and 25 in the x,y and x,z planes. Scale bar: 2 μm g, Lateral point-spread function full-width at half-maximum diameters for each light bead. Mean value shown by horizontal black line. Error bars denote the 95% confidence interval values for the Gaussian fits used to determine PSF widths. h, Axial point-spread function full-width at half-maximum diameters for each light bead. Mean value shown by horizontal black line. Error bars denote the 95% confidence interval values for the Lorentzian fits used to determine PSF widths. i, j Point-spread function FWHM lateral and axial widths, respectively, for the top (z = 0 μm, bead 30), middle (z = 225 μm, bead 18) and bottom (z = 450 μm, bead 1), of the light bead column as a function of radial position in the FOV.
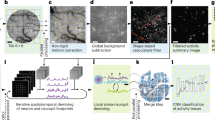
Extended Data Fig. 5 Schematic of the data processing pipeline.
Raw data is assembled into frames and separated into individual temporal stacks for each z plane. Each stack is separately motion-corrected and sent through a constrained non-negative matrix factorization (CNMF) sub-routine to extract neuronal footprints and time-series. Lateral offsets between the planes are accounted for using calibration values, and neurons with both correlated temporal activity and overlapping spatial footprints are merged to prevent doubly-counted cells. Neurons are correlated with vectors representing each stimulus to determine if they are tuned. Example raw and kernel-convolved stimulus vectors and an example time-series for a tuned neuron are shown for uninstructed behaviors during a recording.
Extended Data Fig. 6 Data fidelity and validation in the sparse sampling regime.
a, Extraction fidelity, measured by F-score, as a function of sample spacing. F-score is defined as the harmonic mean of the sensitivity and precision of the neuronal extraction. Solid line indicates mean value, shaded region indicates one standard deviation from the mean. Example images for 0.5, 3, and 5 μm sample spacing inset. b, To-scale schematic of a sparse sampling grid (5 μm sample-to-sample spacing) with a 1 μm FWHM PSF over an example soma. c, 400 example footprints extracted from the data set shown in Fig. 2 with our data processing pipeline. d, Example traces showing the neuropil subtraction mechanism: In-plane signal around each neuronal footprint (left column) is used to estimate the background in the region overlapping the cell body (rightmost column, magenta overlay) and contamination of the extracted transients due to surrounding neuropil. The obtained neuropil signal (magenta lines) is then subtracted from the raw transient signal to result in decontaminated traces (black lines). e, Example traces extracted from a densely sampled (0.5 μm sampling) ground truth data set (red lines) compared to traces extracted from the same data set with intentional down-sampling to reflect the sparse sampling condition (5 μm sampling, blue lines); traces are intentionally offset by 2 × ΔF/F0 for clarity. f, Correlation between traces extracted from 7 ground truth data sets and traces extracted from down-sampled copies of the same data sets, indicating strong correlation (r = 0.91 ± 0.11). g, Comparison of the pixel shifts predicted by our motion correction algorithm for an example ground truth data set (red line) and the same data set after intentional down-sampling (blue line); traces are intentionally offset by 1 μm for clarity. Pixel shift estimates are not affected by the down-sampling.
Extended Data Fig. 7 Normalized heatmaps of extracted neuronal activity.
a, Heatmap of 207,030 neurons extracted from the data set shown in Fig. 2a recorded in a 3 × 5 × 0.5 mm FOV at 4.7 Hz. b, A high resolution subset of 3,000 neurons from the population shown in a. c, Heatmap of 1,065,289 neurons extracted from the data set shown in Fig. 5a recorded in a 5.4 × 6 × 0.5 mm FOV at 2.2. d, A high resolution subset of 3,000 neurons from the population shown in c.
Extended Data Fig. 8 Indicator and extraction statistics.
a, Summary of the number of neurons extracted from 12 recordings with ~3 × 5 × 0.5 mm FOV across N = 6 animals; solid black line denotes the mean, shaded region denotes 1 standard deviation from the mean; data set 1 corresponds to the recording shown in Figs. 2 and 3. b,c,d,e,f Distribution of maximum ΔF/F0 values, baseline noise levels, Z-scores, and transient decay times for transients measured in mice expressing GCaMP6s from the experiment shown in Fig. 2. g, Summary of the number of neurons extracted from 3 recordings with ~5.4 × 6 × 0.5 mm FOV across N = 3 animals; solid black line denotes the mean, shaded region denotes 1 standard deviation from the mean; dataset 1 corresponds to the recording shown in Fig. 5a. h,i,j,k Distributions quantifying neuronal activity for transients extracted from Fig. 5a, following those in b-f.
Extended Data Fig. 9 Hierarchical clustering and trial-to-trial variability analysis of stimulus-tuned neurons from the dataset shown in Fig. 2.
a–c, Correlation distributions of neurons with whisker stimuli, visual stimuli, and uninstructed spontaneous animal behaviors (blue), compared to time-shuffled distributions (red). d, Correlation matrix of all neurons tuned to any stimulus condition. The matrix is sorted by stimulus (boundaries denoted by black lines), cluster, and mean Pearson correlation. e–i, Axial spatial distributions of neurons in clusters 1 through 4 and the uncorrelated population, respectively. j, Distribution of the correlation between trial-to-trial responses of pairs of whisker-tuned neurons compared to a distribution where trial order was randomly shuffled. k, Equivalent distributions to those shown in j for visual-tuned neurons. l, Cumulative fraction of significantly covarying (R > 3σ, Pearson correlation) pairs of neurons as a function of neuron-to-neuron separation. m, Cumulative fraction of significantly covarying (R > 3σ, Pearson correlation) pairs of neurons as a function of neuron-to-neuron axial separation.
Extended Data Fig. 10 Brain heating experimental data and simulations.
a-e, Representative images of brain sections showing immunolabeling for astrocyte activation marker (anti-GFAP, red) and DNA stain (Hoechst 33342, blue) after exposure to the laser power and FOV listed below. Scale bars: 1 mm a, control, no laser exposure. b, 360 mW, 0.4 × 0.4 × 0.5 mm FOV (2250 mW/mm2). c, 250 mW, 3 × 5 × 0.5 mm FOV (17 mW/mm2). d, 450 mW, 3 × 5 × 0.5 mm FOV (34 mW/mm2). e, 250 mW, 0.6 × 0.6 × 0.5 mm FOV (700 mW/mm2). f, Intensity of immunolabeling corresponding to imaging intensity as a fraction compared to mean of control samples. N = 3 separate brain hemispheres per condition. Shaded area denotes the 95% confidence interval of the control group mean. g,h, Simulations of brain temperature at steady state for 450 mW of optical power in a (g) 0.4 mm FOV and a (h) 4 mm FOV; magenta lines indicated nominal focal plane; scale bars: 1 mm. Brain temperature in g heats to ~6 °C above core temperature (37 °C), while heating in h is only ~1 °C. Solid black lines denote boundaries of the cranial window, magenta lines indicate focal plane, dashed black lines indicate contours separated by 2 °C. Scale bar: 1 mm. i, Maximum brain temperature as a function of optical power and FOV. Cooling of the brain through the cranial window leads to a minimum power threshold before the onset of heating in the brain. j, Brain temperature for a fixed power level (250 mW) as a function of cranial window diameter and FOV. Larger cranial windows lead to less overall heating of the brain.
Supplementary information
Supplementary Information
Supplementary Tables 1 and 2 and Supplementary Notes 1–5
Supplementary Video 1
Example mouse behavior during recording. Motion tracking of the right and left paws and right ankle are shown with green, blue, and red dots, respectively.
Supplementary Video 2
3D rendering of the top 56,954 most active neurons from 207,030 detected neurons within a recording volume of ~3 × 5 × 0.5 mm recorded at 4.7 Hz from the data set shown in Fig. 2. Each sphere represents the activity of one neuron, normalized to maximum ΔF/F0 for increased visibility. Increase in opacity and transition from blue to yellow color (following ‘parula’ colormap) indicate increasing calcium activity. Occurrences of whisker stimuli, visual stimuli, or simultaneous whisker and visual stimuli are denoted by the red mark in the upper left-hand corner of the frame. Playback sped up 4×.
Supplementary Video 3
Example recording of a single plane (depth = 344 μm) from the volumetric recording shown in Fig. 2 (3 × 5 mm FOV recorded at 4.7 Hz). Playback sped up 4×. Scale bar, 250 mm.
Supplementary Video 4
Side-by-side comparison of the data set shown in Supplementary Video 3 with (right side) and without (left side) 5 frame averaging.
Supplementary Video 5
Summary of all traces extracted from the recording in Fig. 2.
Supplementary Video 6
Time-lapse of activity onset for neurons tuned to uninstructed behaviors of the animal following the analysis shown in Figs. 3r and 3s. Playback is displayed in real time.
Supplementary Video 7
Example recording of a single plane (depth = 384 μm) from the volumetric recording shown in Figs. 4a–4c (0.6 × 0.6 mm FOV recorded at 9.6 Hz). Playback sped up 4×. Scale bar: 50 mm.
Supplementary Video 8
3D rendering of the top 32,915 most active neurons from 70,275 detected neurons within a recording volume of ~2 × 2 × 0.5 mm recorded at 6.5 Hz from the data set shown in Figs. 4d–4g. Each sphere represents the activity of one neuron, normalized to maximum ΔF/F0 for increased visibility. Increase in opacity and transition from blue to yellow color (following ‘parula’ colormap) indicate increasing calcium activity. Occurrences of whisker stimuli are denoted by the red mark in the upper left-hand corner of the frame. Playback sped up 4×.
Supplementary Video 9
Example recording of a single plane (depth = 144 μm) from the volumetric recording shown in Figs. 4d–4g (2 × 2 mm FOV recorded at 6.5 Hz). Playback sped up 4×. Scale bar: 200 mm.
Supplementary Video 10
3D rendering of the top 150,000 most active neurons from 1,065,289 detected neurons within a recording volume of ~5.4 × 6 × 0.5 mm recorded at 2.2 Hz from the data set shown in Fig. 5a. Each sphere represents the activity of one neuron, normalized to maximum ΔF/F0 for increased visibility. Increase in opacity and transition from blue to yellow color (following ‘parula’ colormap) indicate increasing calcium activity. Playback sped up 4×.
Supplementary Video 11
Example recording of a single plane (depth = 600 μm) from the volumetric recording shown in Fig. 5 (5.4 × 6 mm FOV recorded at 2.2 Hz). Playback sped up 4×. Scale bar: 500 μm.
Supplementary Video 12
Summary of all traces extracted from the recording in Fig. 5.
Source data
Source Data Fig. 2
Source data for all plots within the figure
Source Data Fig. 3
Source data for all plots within the figure
Source Data Fig. 4
Source data for all plots within the figure
Source Data Fig. 5
Source data for all plots within the figure
Source Data Extended Data Fig. 3
Source data for all plots within the figure
Source Data Extended Data Fig. 4
Source data for all plots within the figure
Source Data Extended Data Fig. 6
Source data for all plots within the figure
Source Data Extended Data Fig. 7
Source data for all plots within the figure
Source Data Extended Data Fig. 8
Source data for all plots within the figure
Source Data Extended Data Fig. 10
Source data for all plots within the figure
Rights and permissions
About this article
Cite this article
Demas, J., Manley, J., Tejera, F. et al. High-speed, cortex-wide volumetric recording of neuroactivity at cellular resolution using light beads microscopy. Nat Methods 18, 1103–1111 (2021). https://doi.org/10.1038/s41592-021-01239-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-021-01239-8
This article is cited by
-
Mesoscale volumetric light-field (MesoLF) imaging of neuroactivity across cortical areas at 18 Hz
Nature Methods (2023)
-
Real-time denoising enables high-sensitivity fluorescence time-lapse imaging beyond the shot-noise limit
Nature Biotechnology (2023)
-
Multifocal fluorescence video-rate imaging of centimetre-wide arbitrarily shaped brain surfaces at micrometric resolution
Nature Biomedical Engineering (2023)
-
High-speed low-light in vivo two-photon voltage imaging of large neuronal populations
Nature Methods (2023)
-
FIOLA: an accelerated pipeline for fluorescence imaging online analysis
Nature Methods (2023)