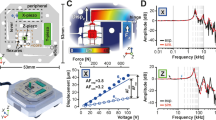
Abstract
Single-molecule Förster resonance energy transfer (smFRET) has become a versatile and widespread method to probe nanoscale conformation and dynamics. However, current experimental modalities often resort to molecule immobilization for long observation times and do not always approach the resolution limit of FRET-based nanoscale metrology. Here we present ABEL-FRET, an immobilization-free platform for smFRET measurements with ultrahigh resolving power in FRET efficiency. Importantly, single-molecule diffusivity is used to provide additional size and shape information for hydrodynamic profiling of individual molecules, which, together with the concurrently measured intramolecular conformation through FRET, enables a holistic and dynamic view of biomolecules and their complexes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
All single-molecule raw data analyzed in this work are available at https://doi.org/10.5281/zenodo.4716779. Source data are provided with this paper.
Code availability
Instrument control and analysis codes used to collect and analyze the data during the current study are supplied as Supplementary Software.
References
Ha, T. et al. Probing the interaction between two single molecules: fluorescence resonance energy transfer between a single donor and a single acceptor. Proc. Natl Acad. Sci. USA 93, 6264–6268 (1996).
Lerner, E. et al. Toward dynamic structural biology: two decades of single-molecule Förster resonance energy transfer. Science 359, eaan1133 (2018).
Rasnik, I., McKinney, S. A. & Ha, T. Surfaces and orientations: much to FRET about? Acc. Chem. Res. 38, 542–548 (2005).
Friedel, M., Baumketner, A. & Shea, J. E. Effects of surface tethering on protein folding mechanisms. Proc. Natl Acad. Sci. USA 103, 8396–8401 (2006).
Cohen, A. E. & Moerner, W. E. Method for trapping and manipulating nanoscale objects in solution. Appl. Phys. Lett. 86, 093109 (2005).
Dienerowitz, M., Howard, J. A. L., Quinn, S. D., Dienerowitz, F. & Leake, M. C. Single-molecule FRET dynamics of molecular motors in an ABEL trap. Methods https://doi.org/10.1016/j.ymeth.2021.01.012 (2021).
Heitkamp, T. & Börsch, M. Fast ATP-dependent subunit rotation in reconstituted FoF1-ATP synthase trapped in solution. Preprint at https://arxiv.org/abs/2103.11986 (2021).
Wang, Q. Enabling Multivariate Investigation of Single-molecule Dynamics in Solution by Counteracting Brownian Motion (Stanford University, 2014); https://searchworks.stanford.edu/view/10689150
Wang, Q. & Moerner, W. E. Single-molecule motions enable direct visualization of biomolecular interactions in solution. Nat. Methods 11, 555–558 (2014).
Watkins, L. P. & Yang, H. Information bounds and optimal analysis of dynamic single molecule measurements. Biophys. J. 86, 4015–4029 (2004).
Holden, S. J. et al. Defining the limits of single-molecule FRET resolution in TIRF microscopy. Biophys. J. 99, 3102–3111 (2010).
Nir, E. et al. Shot-noise limited single-molecule FRET histograms: comparison between theory and experiments. J. Phys. Chem. B 110, 22103–22124 (2006).
Juette, M. F. et al. Single-molecule imaging of non-equilibrium molecular ensembles on the millisecond timescale. Nat. Methods 13, 341–344 (2016).
Tyagi, S. et al. Continuous throughput and long-term observation of single-molecule FRET without immobilization. Nat. Methods 11, 297–300 (2014).
Hanson, J. A. et al. Illuminating the mechanistic roles of enzyme conformational dynamics. Proc. Natl Acad. Sci. USA 104, 18055–18060 (2007).
McKinney, S. A., Déclais, A.-C., Lilley, D. M. J. & Ha, T. Structural dynamics of individual Holliday junctions. Nat. Struct. Biol. 10, 93–97 (2003).
Roy, R., Kozlov, A. G., Lohman, T. M. & Ha, T. SSB protein diffusion on single-stranded DNA stimulates RecA filament formation. Nature 461, 1092–1097 (2009).
Joo, C. et al. Real-time observation of RecA filament dynamics with single monomer resolution. Cell 126, 515–527 (2006).
Cristóvão, M. et al. Single-molecule multiparameter fluorescence spectroscopy reveals directional MutS binding to mismatched bases in DNA. Nucleic Acids Res. 40, 5448–5464 (2012).
Lilley, D. M. J. Structures of helical junctions in nucleic acids. Q. Rev. Biophys. 33, 109–159 (2000).
Roy, R., Kozlov, A. G., Lohman, T. M. & Ha, T. Dynamic structural rearrangements between DNA binding modes of E. coli SSB protein. J. Mol. Biol. 369, 1244–1257 (2007).
Fijen, C. et al. A single-molecule FRET sensor for monitoring DNA synthesis in real time. Phys. Chem. Chem. Phys. 52, 541–553 (2017).
Christian, T. D., Romano, L. J. & Rueda, D. Single-molecule measurements of synthesis by DNA polymerase with base-pair resolution. Proc. Natl Acad. Sci. USA 106, 21109–21114 (2009).
Berezhna, S. Y., Gill, J. P., Lamichhane, R. & Millar, D. P. Single-molecule Förster resonance energy transfer reveals an innate fidelity checkpoint in DNA polymerase I. J. Am. Chem. Soc. 134, 11261–11268 (2012).
Kapanidis, A. N. et al. Alternating-laser excitation of single molecules. Acc. Chem. Res. 38, 523–533 (2005).
Hendrix, J. & Lamb, D. C. Pulsed interleaved excitation: principles and applications. Methods Enzymol. 518, 205–243.
Sonnenfeld, A., Haran, G. & Boukobza, E. Immobilization in surface-tethered lipid vesicles as a new tool for single biomolecule spectroscopy. J. Phys. Chem. B 105, 12165–12170 (2001).
Zhuang, X. et al. Correlating structural dynamics and function in single ribozyme molecules. Science 296, 1473–1476 (2002).
Hellenkamp, B. et al. Precision and accuracy of single-molecule FRET measurements—a multi-laboratory benchmark study. Nat. Methods 15, 669–676 (2018).
Lerner, E. et al. FRET-based dynamic structural biology: challenges, perspectives and an appeal for open-science practices. eLife 10, e60416 (2021).
Wang, Q. & Moerner, W. E. An adaptive anti-Brownian electrokinetic trap with real-time information on single-molecule diffusivity and mobility. ACS Nano 5, 5792–5799 (2011).
Wang, Q. & Moerner, W. E. Optimal strategy for trapping single fluorescent molecules in solution using the ABEL trap. Appl. Phys. B 99, 23–30 (2010).
Ranka, J. K., Windeler, R. S. & Stentz, A. J. Visible continuum generation in air–silica microstructure optical fibers with anomalous dispersion at 800 nm. Opt. Lett. 25, 25 (2000).
Wang, Q. & Moerner, W. E. Lifetime and spectrally resolved characterization of the photodynamics of single fluorophores in solution using the anti-Brownian electrokinetic trap. J. Phys. Chem. B 117, 4641–4648 (2013).
Cohen, A. E. & Moerner, W. E. Controlling Brownian motion of single protein molecules and single fluorophores in aqueous buffer. Opt. Express 16, 6941 (2008).
Wood, S. L. et al. Revealing conformational variants of solution-phase intrinsically disordered tau protein at the single-molecule level. Angew. Chem. Int. Ed. 56, 15584–15588 (2017).
Witucki, G. L. A silane primer: chemistry and applications of aikoxy silanes. J. Coat. Technol. 65, 57–60 (1993).
Watkins, L. P. & Yang, H. Detection of intensity change points in time-resolved single-molecule measurements. J. Phys. Chem. B 109, 617–628 (2005).
Roy, R., Hohng, S. & Ha, T. A practical guide to single-molecule FRET. Nat. Methods 5, 507–516 (2008).
Li, H. & Yang, H. Statistical learning of discrete states in time series. J. Phys. Chem. B 123, 689–701 (2019).
Kim, H. D. et al. Mg2+-dependent conformational change of cRNA studied by fluorescence correlation and FRET on immobilized single molecules. Proc. Natl Acad. Sci. USA 99, 4284–4289 (2002).
Laurence, T. A., Fore, S. & Huser, T. Fast, flexible algorithm for calculating photon correlations. Opt. Lett. 31, 829 (2006).
Sorokina, M., Koh, H. R., Patel, S. S. & Ha, T. Fluorescent lifetime trajectories of a single fluorophore reveal reaction intermediates during transcription initiation. J. Am. Chem. Soc. 131, 9630–9631 (2009).
Sanborn, M. E., Connolly, B. K., Gurunathan, K. & Levitus, M. Fluorescence properties and photophysics of the sulfoindocyanine Cy3 linked covalently to DNA. J. Phys. Chem. B 111, 11064–11074 (2007).
Stennett, E. M. S., Ciuba, M. A., Lin, S. & Levitus, M. Demystifying PIFE: the photophysics behind the protein-induced fluorescence enhancement phenomenon in Cy3. J. Phys. Chem. Lett. 6, 1819–1823 (2015).
Acknowledgements
We thank H. Li for preparation of the adenylate kinase sample, M. Lee for modeling the hydrodynamic property of the HJ, H. Yang for valuable feedback, J. Shaevitz for comments on an early version of the manuscript, E. Gatzogiannis for the loan of a Picoharp 300 unit, R. Windeler (OFS Fitel) for the generous gift of the photonic crystal fiber patch and the staff at Princeton’s PRISM clean room (E. Mills, R. Akhmechet, Z. Lewicka and D. Barth) for assistance with ABEL trap fabrication. We also thank the laboratories of A. Amodeo and S. Shvartsman for access to their gel imager. This work was supported by the Lewis–Sigler Fellowship of Princeton University and US Department of Energy Office of Basic Energy Sciences and Photosynthetic Systems Grant DE-SC0002423.
Author information
Authors and Affiliations
Contributions
H.W. and Q.W. designed the research, performed experiments and analyzed the data. H.W. wrote an initial draft of the manuscript. H.W. and Q.W. discussed and interpreted results and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Methods thanks Jens Michaelis and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Rita Strack was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
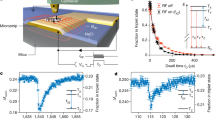
Extended Data Fig. 1 Characterizing EFRET histogram width of end-labeled 11 bp dsDNA on the ABEL-FRET platform.
a, Scheme: we divided a single trapping event into batches of N consecutive photons, calculated the FRET efficiency of each batch and constructed a histogram from data pooled from all measured molecules (a total of 226 molecules). We fit the resultant histogram using a Gaussian distribution and extracted the standard deviation (σ) to quantify histogram width. The inset shows example histograms constructed for N = 750 (σ = 0.024) and N = 6000 (σ = 0.0095). b, FRET histograms of the 11 bp dsDNA sample for N = 200, 2500, 10000 and 20000. c, Histogram width as a function of N for data pooled from all (226) molecules (black symbol) and for one single molecule (magenta symbol). Red symbols represent the simulated shot-noise limit. Parameters used in the simulation were extracted from the 11 bp experiment: E = 0.69, kD,s + kA,s = 13700 s−1, kD,b = 1450 s−1, kA,b = 238 s−1. See Supplementary Note 2 for the definition of these parameters. Red dashed line denotes a linear fit to the simulated shot-noise limit. d, Comparing ABEL-FRET with other smFRET modalities. Integration time (top axis) is calculated from N using a measured detection rate of 15.4 kHz. Symbols mark literature values by different smFRET modalities. (Nir et al.: Ref. 12; Holden et al.: Ref. 11; Juette et al.: Ref. 13; Tyagi et al. Ref. 14) Vertical errorbars in the ABEL-FRET dataset represent 95% confidence intervals for the fitted standard deviation (σ). We emphasize that the near shot-noise limited width in panel C, was obtained using the Cy3-Cy5 FRET pair (Supplementary Table 3). Other popular FRET pairs (for example Alexa488-Alexa594, Cy3B-Atto647N, etc) have yet to be characterized.
Extended Data Fig. 2 ABEL-FRET achieves narrow smFRET histograms on the model protein adenylate kinase (AK).
a, Structure and FRET labeling of E. Coli AK (PDB:1AKE). b, Sample validation: a single band consistent with the molecular weight of AK (~26 kDa) was observed on SDS-PAGE gel with fluorescence signal in both Cy3 and Cy5 channels. The gel images were obtained from a single experiment. c-e, Example ABEL-FRET traces of AK-Alexa555-Alexa647 without substrate. Most molecules show D values around ~85 µm2/s, consistent with hydrodynamic modeling based on the crystal structure (Supplementary Note 7). About 4% of the molecules show much lower D values (< 60 μm2/s, not shown). These were considered oligomers and excluded from further analysis even though FRET was observed. f, An example AK molecule in the presence of 0.5 mM AMP-PNP (adenosine 5’-[β,γ-imido]triphosphate, a nonhydrolyzable ATP analog) and 0.5 mM AMP showing frequent millisecond dynamics, similar to previously observed15. Note the intensity binning time is 2 ms in this example. g, Two-dimensional (D-EFRET) distribution of 296 AK molecules measured without substrate and the corresponding FRET histogram. Each symbol represents D and EFRET values averaged over a single AK molecule. Among these molecules, a mean of 16,200 photons were detected, lower than the experiment on Cy3-Cy5 labeled DNA (25,200 photons, Fig. 1c). h, The FRET histogram width σ(EFRET) as a function of N for four single molecules (solid colored lines) and data pooled from 296 AK molecules (magenta circles). Data on 11 bp DNA (gray circles) and the corresponding shot-noise limit (red dashed line) are plotted as references. The horizontal dashed line (σ = 0.026) represents the width extracted from the molecule-by-molecule analysis in panel G. In this case, the single-molecule measurements were close to shot-noise limited but combining different molecules resulted in broadening in excess of the shot-noise limit. This is strong indication that molecule-to-molecule heterogeneity in this sample is responsible for the extra broadening.
Extended Data Fig. 3 Validating the population identification of the 13-component DNA mixture experiment.
Top: FRET histogram of the 13-component dsDNA experiment with a 12-component Gaussian fit. The mean and standard deviation of each population were extracted from the fit and tabulated. Note that the 10 bp DNA was included in the mixture but not well resolved (arrow), likely due to dissociation at the ~pM concentration in the experiment. Bottom: individual measurements containing a specific subset of the mixture (see figure legend for details, each color represents one experiment). Dashed lines indicate good alignment between the samples. In the 13-component mixture experiment, it took a total of 283 minutes to acquire 1477 FRET-active molecules. The total data acquisition time includes molecule-by-molecule smFRET recording, waiting for new molecules to diffuse into the trapping region and measuring donor only molecules.
Extended Data Fig. 4 Kinetic rates of Holliday junction dynamics.
a, Donor-acceptor intensity cross-correlation curves of HJ for 1 mM Mg2+ (orange, averaged over 44 molecules) and 5 mM Mg2+ (blue, averaged over 59 molecules). Solid lines are single exponential fits. b, Extracted rates of HJ dynamics and comparison to literature values. Data are presented as fitted rates (ktot) +/− 95% C.I. from the fits. c, Single-molecule rates of HJ dynamics. Example fits from two molecules (red and blue circles) are shown on the top for each condition (normalized to the peak amplitudes). In the scatter plots, data along the ktot axis are presented as fitted rates + /− 95% C.I. from the fits. Further investigations are needed to determine if these apparent differences in ktot are due to molecule-to-molecule heterogeneity or insufficient sampling of relatively short time traces.
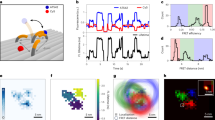
Extended Data Fig. 5 ABEL-FRET monitors slow (~1 s−1) dynamics of the RecA-ssDNA nucleofilament.
a, Top schematic: The experimental design is based on the smFRET assay developed in Ref. 18. The DNA sample is a partial duplex with a 60nt ssDNA overhang ((dT)10+49). In the presence of ATP, the RecA protein subunits (blue ovals) are expected to bind the ssDNA overhang to form a nucleofilament that undergoes binding/unbinding dynamics near the dsDNA-ssDNA junction (that is the 5’-end of the ssDNA region). The FRET dyes are installed at the 5’-end of the ssDNA overhang to monitor distance changes induced by binding and unbinding of RecA protein. The separation between the FRET dyes is kept short (here 10 nt) to allow binding of only a few (here 2) RecA monomers between them. Additional RecA monomers cannot bind if the ssDNA region between the FRET pair has reached maximum occupancy (two monomers). Colored stars mark the positions of the FRET dyes. Blue ovals represent RecA monomers. Dark blue ovals mark RecA monomers that are bound in between the FRET dyes. Bottom: Example time traces under the conditions marked above each panel. We observed three major FRET states (M0 ~ 0.87, M1 ~ 0.58, M2 ~ 0.35, horizontal dashed lines) and transitions between the three. These observations are similar to TIRF microscopy measurements on immobilized DNA substrates18 and were previously interpreted to represent RecA protein binding and unbinding. We also observed many transient (<100 ms) states (arrows) which could be missed in previous TIRFM measurements due to limited time resolution. b, Histogram of observed FRET levels, showing three major populations at M0, M1 and M2 and at least one possible minor state (arrow) at ~0.45. Notably, our FRET efficiency histogram is much sharper compared to that obtained on immobilized molecules18. c, (dT)10+49 in the presence of 1 µM RecA and 1 mM ATPγS only shows the M2 state with no dynamics, representing the fully formed nucleofilament. This observation also agrees with previous experiments on immobilized DNA substrates18.
Extended Data Fig. 6 More example ABEL-FRET traces that visualize the DNA damage-repair cycle.
a, Substrate DNA only. b, Damaged DNA only. c, Substrate DNA incubated with nicking endonuclease. d-j, Substrate DNA with all components of the damage-repair cycle. Protein binding events were visualized by transient drops of diffusion coefficient. Three types of binding events were observed and annotated: binding that changes the EFRET of DNA from low to high was identified to be nicking endonuclease damaging the DNA (orange squares), binding that changes the EFRET of DNA from high to low was identified to be the polymerase repairing the damaged DNA (blue circles), binding that does not change EFRET was considered non-productive (green plus signs). FRET fluctuations during polymerization events were frequently observed (shaded areas in panels D, H and I). Panel H shows an example trace that needed two consecutive (partial) polymerization runs (from 3–5.5 seconds, events marked with two blue circles) to completely repair. k, Mapping of observed binding events categorized by the structural consequences they induce on the substrate DNA.
Extended Data Fig. 7 ABEL-FRET incorporates excited-state lifetime measurements.
a-d, Example single-molecule traces showing intensity, feedback voltages, FRET efficiency and photon delay time histograms with fits. The sample is a mixture of 11 bp, 12 bp and 13 bp DNA duplexes. e, 2D scatter plot of corrected FRET efficiency versus donor lifetime. Black dots represent data acquired from single molecules. The underlying density is visualized using a 2D kernel density estimation algorithm. A total of 92 molecules were measured. The dotted red line shows the theoretical relationship E = 1 - τ/τ0, where the value of τ0 is extracted from the mean lifetime of the donor only species. f, EFRET values calculated from intensity and lifetime-based measurements agree well. Background, donor leakage and γ corrections were applied to the intensity-based FRET calculation. G, Our measured lifetime of Cy3 on dsDNA is consistent with literature values. Sorokina et al.: Ref. 43, Sanborn et al.: Ref. 44, Stennett et al.: Ref. 45.
Supplementary information
Supplementary Information
Supplementary Figs. 1–11, Tables 1–4, Notes 1–8, Descriptions for Supplementary Software 1 and 2 and Bibliography.
Supplementary Software 1
ABEL-FRET instrument control software (SMTrapV4).
Supplementary Software 2
Graphical User Interface (TrapViewV4) for visualization and analysis of ABEL-FRET data.
Source data
Source Data Extended Data Fig. 2
Source data and raw gel images.
Rights and permissions
About this article
Cite this article
Wilson, H., Wang, Q. ABEL-FRET: tether-free single-molecule FRET with hydrodynamic profiling. Nat Methods 18, 816–820 (2021). https://doi.org/10.1038/s41592-021-01173-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-021-01173-9
This article is cited by
-
Stable trapping of multiple proteins at physiological conditions using nanoscale chambers with macromolecular gates
Nature Communications (2023)