Abstract
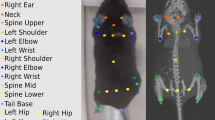
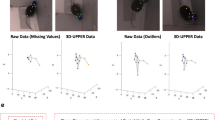
We developed a three-dimensional (3D) synthetic animated mouse based on computed tomography scans that is actuated using animation and semirandom, joint-constrained movements to generate synthetic behavioral data with ground-truth label locations. Image-domain translation produced realistic synthetic videos used to train two-dimensional (2D) and 3D pose estimation models with accuracy similar to typical manual training datasets. The outputs from the 3D model-based pose estimation yielded better definition of behavioral clusters than 2D videos and may facilitate automated ethological classification.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
All data and raw behavioral video images are available online at Open Science Framework: https://osf.io/h3ec5/ (ref. 2). Source data for Fig. 2 and Supplementary Figs. 1–3 are provided with this paper.
Code availability
All code and models are available on public repositories: https://github.com/ubcbraincircuits/mCBF (ref. 13) and https://osf.io/h3ec5/ (ref. 2); see workflow document for an overview of software steps. Code (KID score, U-GAT-IT, pose lifting and clustering) is set up for reproducible runs via a capsule on Code Ocean23.
References
Mathis, A. et al. DeepLabCut: markerless pose estimation of user-defined body parts with deep learning. Nat. Neurosci. 21, 1281–1289 (2018).
Bolanos, L. et al. Synthetic animated mouse (SAM), University of British Columbia, data and 3D-models. OSF https://doi.org/10.17605/OSF.IO/H3EC5 (2021).
Khmelinskii, A. et al. Articulated whole-body atlases for small animal image analysis: construction and applications. Mol. Imaging Biol. 13, 898–910 (2011).
Klyuzhin, I. S., Stortz, G. & Sossi, V. Development of a digital unrestrained mouse phantom with non-periodic deformable motion. In Proc. 2015 IEEE Nuclear Science Symposium and Medical Imaging Conference (NSS/MIC) (IEEE, 2015); https://doi.org/10.1109/nssmic.2015.7582140
Li, S. et al. Deformation-aware unpaired image translation for pose estimation on laboratory animals. 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) https://doi.org/10.1109/cvpr42600.2020.01317 (IEEE, 2020).
Radhakrishnan, S. & Kuo, C. J. Synthetic to real world image translation using generative adversarial networks. IEEE Conference Publication https://ieeexplore.ieee.org/stamp/stamp.jsp?tp=&arnumber=8493745 (IEEE, 2018).
The International Brain Laboratory et al. Standardized and reproducible measurement of decision-making in mice. Preprint at bioRXiv https://doi.org/10.1101/2020.01.17.909838 (2020).
Galiñanes, G. L., Bonardi, C. & Huber, D. Directional reaching for water as a cortex-dependent behavioral framework for mice. Cell Rep. 22, 2767–2783 (2018).
Kwak, I. S., Guo, J.-Z., Hantman, A., Kriegman, D. & Branson, K. Detecting the starting frame of actions in video. Preprint at https://arxiv.org/abs/1906.03340 (2019).
Kim, J., Kim, M., Kang, H. & Lee, K. U-GAT-IT: generative attentional networks with adaptive layer-instance normalization for image-to-image translation. Preprint at https://arxiv.org/abs/1907.10830 (2019).
Wang, Z., Simoncelli, E. P. & Bovik, A. C. Multiscale structural similarity for image quality assessment. In Proc. Thirty-Seventh Asilomar Conference on Signals, Systems Computers, 2003 Vol. 2 1398–1402 (2003).
Bińkowski, M., Sutherland, D. J., Arbel, M. & Gretton, A. Demystifying MMD GANs. Preprint at https://arxiv.org/abs/1801.01401 (2018).
Bolanos, L. et al. ubcbraincircuits/mCBF: mCBF. Zenodo https://doi.org/10.5281/zenodo.4563193 (2021).
Nath, T. et al. Using DeepLabCut for 3D markerless pose estimation across species and behaviors. Nat. Protoc. 14, 2152–2176 (2019).
Martinez, J., Hossain, R., Romero, J. & Little, J. J. A simple yet effective baseline for 3D human pose estimation. In Proc. 2017 IEEE International Conference on Computer Vision (ICCV) (IEEE, 2017); https://doi.org/10.1109/iccv.2017.288
Günel, S. et al. DeepFly3D, a deep learning-based approach for 3D limb and appendage tracking in tethered, adult Drosophila.eLife 8, e48571 (2019).
Hopkins, B. & Skellam, J. G. A new method for determining the type of distribution of plant individuals. Ann. Bot. 18, 213–227 (1954).
Levine, J. H. et al. Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell 162, 184–197 (2015).
Blondel, V. D., Guillaume, J.-L., Lambiotte, R. & Lefebvre, E. Fast unfolding of communities in large networks. J. Stat. Mech. 2008, P10008 (2008).
Vogt, N. Collaborative neuroscience. Nat. Methods 17, 22 (2020).
Wang, Z., Bovik, A. C., Sheikh, H. R. & Simoncelli, E. P. Image quality assessment: from error visibility to structural similarity. IEEE Trans. Image Process. 13, 600–612 (2004).
Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J. & Wojna, Z. Rethinking the inception architecture for computer vision. In Proc. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) (IEEE, 2016); https://doi.org/10.1109/cvpr.2016.308.
Bolaños, L. A. et al. A 3D virtual mouse generates synthetic training data for behavioral analysis. Code Ocean https://doi.org/10.24433/CO.5412865.v1 (2021).
Acknowledgements
This work was supported by a Canadian Institutes of Health Research (CIHR) FDN-143209, NIH R21, a Fondation Leducq grant, Brain Canada for the Canadian Neurophotonics Platform and a Canadian Partnership for Stroke Recovery Catalyst grant to T.H.M. H.R. is supported by grants from Natural Sciences and Engineering Research Council of Canada (NSERC). D.X. was supported in part by funding provided by Brain Canada, in partnership with Health Canada, for the Canadian Open Neuroscience Platform initiative. This work was supported by computational resources made available through the NeuroImaging and NeuroComputation Centre at the Djavad Mowafaghian Centre for Brain Health (RRID SCR_019086). N.L.F. is supported by an NSERC Discovery Grant. Micro-CT imaging was performed at the UBC Centre for High-Throughput Phenogenomics, a facility supported by the Canada Foundation for Innovation, British Columbia Knowledge Development Foundation and the UBC Faculty of Dentistry. We thank E. Koch and L. Raymond of the UBC Djavad Mowafaghian Centre for Brain Health for the mouse open field video.
Author information
Authors and Affiliations
Contributions
L.A.B., T.H.M., J.M.L. and D.X. developed the synthetic mouse concept. L.A.B. generated the mouse model and all main figure animations. C.D. made supplemental models. L.A.B., T.H.M., J.M.L. and D.X. wrote the manuscript. H.R. edited the manuscript and advised on algorithm selection and validation statistics. D.X. collected experimental data and performed data analysis. H.H., L.A.B., D.X. and P.K.G. performed modeling and data analysis. N.L.F. collected the computed tomography scans and advised on model selection and resolution.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Methods thanks the anonymous reviewers for their contribution to the peer review of this work. Nina Vogt was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–3 and Table 1.
Supplementary Video 1
Synthetic data tutorial.
Supplementary Video 2
Mouse model.
Supplementary Video 3
Style transfer U-GAT-IT model (wheel).
Supplementary Video 4
Style transfer U-GAT-IT model (open field).
Supplementary Video 5
Style transfer U-GAT-IT model (Janelia pellet reaching).
Supplementary Video 6
Model gallery.
Supplementary Video 7
Water reaching and grooming scenes.
Supplementary Video 8
DLC 3D pose lifting.
Supplementary Data 1
Source data Supplementary Fig. 1.
Supplementary Data 2
Source data Supplementary Fig. 2.
Supplementary Data 3
Source data Supplementary Fig. 3.
Source data
Source Data Fig. 2
Statistical source data.
Rights and permissions
About this article
Cite this article
Bolaños, L.A., Xiao, D., Ford, N.L. et al. A three-dimensional virtual mouse generates synthetic training data for behavioral analysis. Nat Methods 18, 378–381 (2021). https://doi.org/10.1038/s41592-021-01103-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-021-01103-9
This article is cited by
-
A primary sensory cortical interareal feedforward inhibitory circuit for tacto-visual integration
Nature Communications (2024)
-
replicAnt: a pipeline for generating annotated images of animals in complex environments using Unreal Engine
Nature Communications (2023)
-
Large-scale capture of hidden fluorescent labels for training generalizable markerless motion capture models
Nature Communications (2023)
-
Automated phenotyping of postoperative delirium-like behaviour in mice reveals the therapeutic efficacy of dexmedetomidine
Communications Biology (2023)
-
Three-dimensional surface motion capture of multiple freely moving pigs using MAMMAL
Nature Communications (2023)