Abstract
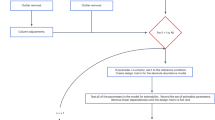
MassIVE.quant is a repository infrastructure and data resource for reproducible quantitative mass spectrometry–based proteomics, which is compatible with all mass spectrometry data acquisition types and computational analysis tools. A branch structure enables MassIVE.quant to systematically store raw experimental data, metadata of the experimental design, scripts of the quantitative analysis workflow, intermediate input and output files, as well as alternative reanalyses of the same dataset.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
All the datasets that support this study are publicly available in MassIVE.quant (https://massive.ucsd.edu/ProteoSAFe/static/massive-quant.jsp) with MassIVE and ProteomeXchange identifiers. Additionally, identifiers for all the datasets are listed in Supplementary Tables 2, 3 and 5.
References
Peng, R. D. Reproducible research in computational science. Science 334, 1226–1227 (2011).
Wilkinson, M. D. et al. The FAIR guiding principles for scientific data management and stewardship. Sci. Data 3, 160018 (2016).
Perez-Riverol, Y. et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47, D442–D450 (2019).
Sharma, V. et al. Panorama: a targeted proteomics knowledge base. J. Proteome Res. 13, 4205–4210 (2014).
Sharma, V. et al. Panorama public: a public repository for quantitative data sets processed in skyline. Mol. Cell Proteom. 17, 1239–1244 (2018).
Farrah, T. et al. PASSEL: the PeptideAtlas SRMexperiment library. Proteomics 12, 1170–1175 (2012).
Vizcaino, J. A. et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 (2014).
Deutsch, E. W. et al. The ProteomeXchange consortium in 2017: supporting the cultural change in proteomics public data deposition. Nucleic Acids Res. 45, D1100–D1106 (2017).
MacLean, B. et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 (2010).
Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008).
Rost, H. L. et al. OpenMS: a flexible open-source software platform for mass spectrometry data analysis. Nat. Methods 13, 741–748 (2016).
Rost, H. L. et al. OpenSWATH enables automated, targeted analysis of data-independent acquisition MS data. Nat. Biotechnol. 32, 219–223 (2014).
Tsou, C. C. et al. DIA-Umpire: comprehensive computational framework for data-independent acquisition proteomics. Nat. Methods 12, 258–264 (2015).
Bruderer, R. et al. Extending the limits of quantitative proteome profiling with data-independent acquisition and application to acetaminophen-treated three-dimensional liver microtissues. Mol. Cell Proteom. 14, 1400–1410 (2015).
Navarro, P. et al. A multicenter study benchmarks software tools for label-free proteome quantification. Nat. Biotechnol. 34, 1130–1136 (2016).
Choi, M. et al. ABRF Proteome informatics research group (iPRG) 2015 study: detection of differentially abundant proteins in label-free quantitative LC-MS/MS experiments. J. Proteome Res. 16, 945–957 (2017).
Choi, M. et al. MSstats: an R package for statistical analysis of quantitative mass spectrometry-based proteomic experiments. Bioinformatics 30, 2524–2526 (2014).
Selevsek, N. et al. Reproducible and consistent quantification of the Saccharomyces cerevisiae proteome by SWATH-mass spectrometry. Mol. Cell Proteom. 14, 739–749 (2015).
Acknowledgements
This work was supported in part by NSF CAREER award no. DBI-1054826, grant no. NSF DBI-1759736 and the Chan-Zuckerberg foundation to O.V., grant no. NIH-NLM 1R01LM013115 to N.B. and O.V., NSF award no. ABI 1759980, NIH award nos. P41GM103484 and R24GM127667 to N.B. and the Personalized Health and Related Technologies (grant no. PHRT 0-21411-18) strategic focus area of ETH to B.W. The CRG/UPF Proteomics Unit is part of the Spanish Infrastructure for Omics Technologies (ICTS OmicsTech) and it is a member of the ProteoRed PRB3 consortium that is supported by grant no. PT17/0019 of the PE I+D+i 2013–2016 from the Instituto de Salud Carlos III (ISCIII) and ERDF. We acknowledge support from the Spanish Ministry of Science, Innovation and Universities, ‘Centro de Excelencia Severo Ochoa 2013–2017’, SEV-2012–0208 and Secretaria d’Universitats i Recerca del Departament d’Economia i Coneixement de la Generalitat de Catalunya (grant no. 2017SGR595). This project has received funding from the European Union’s Horizon 2020 research and innovation program under grant agreement no. 823839 (EPIC-XS). Y.P.-R. acknowledges the Wellcome Trust (grant no. 208391/Z/17/Z). We thank the MacCoss laboratory (Department of Genome Sciences, University of Washington) for the Skyline analyses and contributing the processed data, the Slavov laboratory (College of Engineering, Northeastern University) for providing the data and the Guo laboratory (School of Life Sciences, Westlake University, China) for providing the data.
Author information
Authors and Affiliations
Contributions
M.C., J.C., N.B. and O.V. designed the research. M.C. and T.H. collected datasets and performed statistical analysis. J.C. and B.P. implemented MassIVE.quant. T.-H.T. performed statistical analysis. C.C., E.S. and M.T. experimented with new controlled mixtures. C.C., M.T., R.H., G.C.T., Y.P-R., J.M., M.M., S.G., M.P., E.V., B.W., O.M.B., A.I.N., L.R., T.D. and E.S. analyzed data up to quantification. M.C., N.B. and O.V. wrote the manuscript, with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
O.M.B., J.M. and L.R. are employees of Biognosys AG. Spectronaut is a trademark of Biognosys AG. M.T. and T.D. are employees of Hoffmann-La Roche Ltd. All other authors declare no competing interests.
Additional information
Peer review information Arunima Singh was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Note 1, Figs. 1–4, Tables 1–5 and references.
Rights and permissions
About this article
Cite this article
Choi, M., Carver, J., Chiva, C. et al. MassIVE.quant: a community resource of quantitative mass spectrometry–based proteomics datasets. Nat Methods 17, 981–984 (2020). https://doi.org/10.1038/s41592-020-0955-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-020-0955-0
This article is cited by
-
Size-exclusion chromatography combined with DIA-MS enables deep proteome profiling of extracellular vesicles from melanoma plasma and serum
Cellular and Molecular Life Sciences (2024)
-
Biological big-data sources, problems of storage, computational issues, and applications: a comprehensive review
Knowledge and Information Systems (2024)
-
Extracellular vesicle analysis
Nature Reviews Methods Primers (2023)
-
A proteomic meta-analysis refinement of plasma extracellular vesicles
Scientific Data (2023)
-
Cardinal v.3: a versatile open-source software for mass spectrometry imaging analysis
Nature Methods (2023)