Abstract
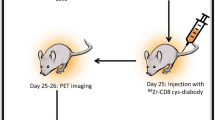
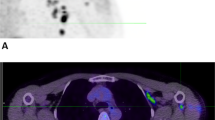
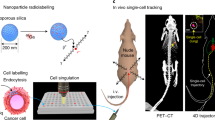
The immune system’s ability to recognize peptides on major histocompatibility molecules contributes to the eradication of cancers and pathogens. Tracking these responses in vivo could help evaluate the efficacy of immune interventions and improve mechanistic understanding of immune responses. For this purpose, we employ synTacs, which are dimeric major histocompatibility molecule scaffolds of defined composition. SynTacs, when labeled with positron-emitting isotopes, can noninvasively image antigen-specific CD8+ T cells in vivo. Using radiolabeled synTacs loaded with the appropriate peptides, we imaged human papillomavirus-specific CD8+ T cells by positron emission tomography in mice bearing human papillomavirus-positive tumors, as well as influenza A virus–specific CD8+ T cells in the lungs of influenza A virus–infected mice. It is thus possible to visualize antigen-specific CD8+ T-cell populations in vivo, which may serve prognostic and diagnostic roles.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The original PET–CT DICOM files that support the findings of this study are available from the corresponding author upon reasonable request. They are not available from public repositories due to their large size (~1 GB per mouse). Source data are provided with this paper.
References
Rashidian, M. et al. Predicting the response to CTLA-4 blockade by longitudinal noninvasive monitoring of CD8+ T cells. J. Exp. Med. 214, 2243–2255 (2017).
Wu, A. M. Antibodies and antimatter: the resurgence of immuno-PET. J. Nucl. Med. 50, 2–5 (2009).
Tavare, R. et al. An effective immuno-PET imaging method to monitor CD8-dependent responses to immunotherapy. Cancer Res. 76, 73–82 (2016).
Dey, S. et al. Tracking antigen-specific T cells: technological advancement and limitations. Biotechnol. Adv. 37, 145–153 (2019).
Alam, I. S. et al. Imaging activated T cells predicts response to cancer vaccines. J. Clin. Invest. 128, 2569–2580 (2018).
Larimer, B. M. et al. Granzyme B PET imaging as a predictive biomarker of immunotherapy response. Cancer Res. 77, 2318–2327 (2017).
Gibson, H. M. et al. IFN-γ PET imaging as a predictive tool for monitoring response to tumor immunotherapy. Cancer Res. 78, 5706–5717 (2018).
Marciscano, A. E. & Thorek, D. L. J. Role of noninvasive molecular imaging in determining response. Adv. Radiat. Oncol. 3, 534–547 (2018).
Vyas, J. M., Van der Veen, A. G. & Ploegh, H. L. The known unknowns of antigen processing and presentation. Nat. Rev. Immunol. 8, 607–618 (2008).
Skeate, J. G., Woodham, A. W., Einstein, M. H., Da Silva, D. M. & Kast, W. M. Current therapeutic vaccination and immunotherapy strategies for HPV-related diseases. Hum. Vaccin. Immunother. 12, 1418–1429 (2016).
Sebzda, E. et al. Selection of the T cell repertoire. Annu. Rev. Immunol. 17, 829–874 (1999).
Davis, M. M. et al. Ligand recognition by αβ T cell receptors. Annu. Rev. Immunol. 16, 523–544 (1998).
Huppa, J. B. & Davis, M. M. T-cell-antigen recognition and the immunological synapse. Nat. Rev. Immunol. 3, 973–983 (2003).
Doherty, P. C. The tetramer transformation. J. Immunol. 187, 5–6 (2011).
Altman, J. D. et al. Phenotypic analysis of antigen-specific T lymphocytes. Science 274, 94–96 (1996).
Duncan, A. R. & Winter, G. The binding site for C1q on IgG. Nature 332, 738–740 (1988).
Wines, B. D., Powell, M. S., Parren, P. W., Barnes, N. & Hogarth, P. M. The IgG Fc contains distinct Fc receptor (FcR) binding sites: the leukocyte receptors Fc γ RI and Fc γ RIIa bind to a region in the Fc distinct from that recognized by neonatal FcR and protein A. J. Immunol. 164, 5313–5318 (2000).
Feltkamp, M. C. et al. Vaccination with cytotoxic T lymphocyte epitope-containing peptide protects against a tumor induced by human papillomavirus type 16-transformed cells. Eur. J. Immunol. 23, 2242–2249 (1993).
Rotzschke, O. et al. Isolation and analysis of naturally processed viral peptides as recognized by cytotoxic T cells. Nature 348, 252–254 (1990).
Gallimore, A. et al. A protective cytotoxic T cell response to a subdominant epitope is influenced by the stability of the MHC class I/peptide complex and the overall spectrum of viral peptides generated within infected cells. Eur. J. Immunol. 28, 3301–3311 (1998).
Rashidian, M. et al. Noninvasive imaging of immune responses. Proc. Natl Acad. Sci. USA 112, 6146–6151 (2015).
Woodham, A. W. et al. Nanobody-antigen conjugates elicit HPV-specific antitumor immune responses. Cancer Immunol. Res. 6, 870–880 (2018).
Rashidian, M. et al. The use of (18)F-2-fluorodeoxyglucose (FDG) to label antibody fragments for immuno-PET of pancreatic cancer. ACS Cent. Sci. 1, 142–147 (2015).
Walboomers, J. M. et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 189, 12–19 (1999).
Stanley, M. A., Pett, M. R. & Coleman, N. HPV: from infection to cancer. Biochem. Soc. Trans. 35, 1456–1460 (2007).
Feltkamp, M. C. et al. Cytotoxic T lymphocytes raised against a subdominant epitope offered as a synthetic peptide eradicate human papillomavirus type 16-induced tumors. Eur. J. Immunol. 25, 2638–2642 (1995).
Kanodia, S. et al. Expression of LIGHT/TNFSF14 combined with vaccination against human papillomavirus Type 16 E7 induces significant tumor regression. Cancer Res. 70, 3955–3964 (2010).
Thomas, P. G., Keating, R., Hulse-Post, D. J. & Doherty, P. C. Cell-mediated protection in influenza infection. Emerg. Infect. Dis. 12, 48–54 (2006).
Wu, A. M. Engineered antibodies for molecular imaging of cancer. Methods 65, 139–147 (2014).
Sheeley, D. M., Merrill, B. M. & Taylor, L. C. Characterization of monoclonal antibody glycosylation: comparison of expression systems and identification of terminal α-linked galactose. Anal. Biochem. 247, 102–110 (1997).
Roggenbuck, D., Mytilinaiou, M. G., Lapin, S. V., Reinhold, D. & Conrad, K. Asialoglycoprotein receptor (ASGPR): a peculiar target of liver-specific autoimmunity. Auto. Immun. Highlights 3, 119–125 (2012).
Pyzik, M. et al. Hepatic FcRn regulates albumin homeostasis and susceptibility to liver injury. Proc. Natl Acad. Sci. USA 114, E2862–E2871 (2017).
Sockolosky, J. T. & Szoka, F. C. The neonatal Fc receptor, FcRn, as a target for drug delivery and therapy. Adv. Drug Deliv. Rev. 91, 109–124 (2015).
Weissleder, R., Schwaiger, M. C., Gambhir, S. S. & Hricak, H. Imaging approaches to optimize molecular therapies. Sci. Transl. Med. 8, 355ps316 (2016).
Mall, S. et al. Immuno-PET imaging of engineered human T cells in tumors. Cancer Res. 76, 4113–4123 (2016).
Seo, J. W. et al. CD8(+) T-cell density imaging with (64)Cu-labeled Cys-diabody informs immunotherapy protocols. Clin. Cancer Res. 24, 4976–4987 (2018).
Quayle, S. N. et al. CUE-101, a novel HPV16 E7-pHLA-IL-2-Fc fusion protein, enhances tumor antigen-specific T cell activation for the treatment of HPV16-driven malignancies. Clin. Cancer Res. 26, 1953–1964 (2020).
Guimaraes, C. P. et al. Site-specific C-terminal and internal loop labeling of proteins using sortase-mediated reactions. Nat. Protoc. 8, 1787–1799 (2013).
Steurer, W. et al. Ex vivo coating of islet cell allografts with murine CTLA4/Fc promotes graft tolerance. J. Immunol. 155, 1165–1174 (1995).
Yan, L., Woodham, A. W., Da Silva, D. M. & Kast, W. M. Functional analysis of HPV-like particle-activated Langerhans cells in vitro. Methods Mol. Biol. 1249, 333–350 (2015).
Carvalho, L. H., Hafalla, J. C. & Zavala, F. ELISPOT assay to measure antigen-specific murine CD8(+) T cell responses. J. Immunol. Methods 252, 207–218 (2001).
Schmidt, F. I. et al. Phenotypic lentivirus screens to identify functional single domain antibodies. Nat. Microbiol. 1, 16080 (2016).
Acknowledgements
A.W.W. is supported by the Arnold O. Beckman Postdoctoral Fellowship. R.W.C. is supported in part by funding from the Cancer Research Institute Irvington Postdoctoral Fellowship. M.R. was supported by the National Institutes of Health (grant 1K22CA226040-01). H.L.P. is supported by the Lustgarten Foundation (award ID 388167). The laboratory of H.L.P. receives financial support in the form of a sponsored research agreement from VIR in the general area of immunity to flu virus. We acknowledge Cue Biopharma for partial support of this work. The synTac technology was developed with support provided by the National Institutes of Health (U01GM094665, U54GM094662, R01AI145024 and R01CA198095 to S.C.A.). We acknowledge the Wollowick Family Foundation Chair in Multiple Sclerosis and Immunology (to S.C.A.) and Janet & Martin Spatz and the Helen & Irving Spatz Foundation. Additional support provided by the Albert Einstein Macromolecular Therapeutics Development Facility, the Einstein-Rockefeller-CUNY Center for AIDS Research (P30AI124414) and the Albert Einstein Cancer Center (P30CA013330).
Author information
Authors and Affiliations
Contributions
A.W.W., S.H.Z., E.L.Z., S.C.A. and H.L.P. conceived of and designed experiments. S.H.Z., R.J.C., R.D.S., S.J.G. and S.C.A. designed and assembled the plasmids encoding the heavy and light chains of the synTacs. A.W.W., E.L.Z. and M.R. performed preliminary HPV, IAV and PET studies, respectively. A.W.W., S.H.Z., E.L.Z., S.C.K., R.W.C., M.R., S.J.G., J.L.D., M.M., P.K.D. and A.B.P. performed experiments. A.W.W., S.H.Z., E.L.Z., S.C.A. and H.L.P. analyzed and interpreted data. A.W.W., S.H.Z., S.C.A. and H.L.P. drafted the manuscript. All authors reviewed and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The synTac technology was developed in the laboratory of S.C.A. and was licensed to Cue Biopharma, Inc., in which he holds equity, receives royalties and serves as chair of its scientific advisory board. S.J.G. and R.D.S. receive royalties from Cue Biopharma, Inc. The laboratory of H.P. receives financial support in the form of a sponsored research agreement from VIR Biotechnology in the general area of immunity to flu virus, but H.L.P. has no equity stake in VIR.
Additional information
Peer review information Nina Vogt was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–14, Tables 1–6 and Source Data for Supplementary Figs. 1 and 3.
Source data
Source Data Fig. 1
Unmodified Coomassie-stained gel; unprocessed western blot.
Source Data Fig. 6
Unmodified Coomassie-stained gels.
Rights and permissions
About this article
Cite this article
Woodham, A.W., Zeigler, S.H., Zeyang, E.L. et al. In vivo detection of antigen-specific CD8+ T cells by immuno-positron emission tomography. Nat Methods 17, 1025–1032 (2020). https://doi.org/10.1038/s41592-020-0934-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-020-0934-5