Abstract
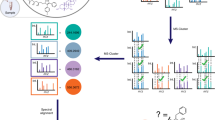
Molecular networking has become a key method to visualize and annotate the chemical space in non-targeted mass spectrometry data. We present feature-based molecular networking (FBMN) as an analysis method in the Global Natural Products Social Molecular Networking (GNPS) infrastructure that builds on chromatographic feature detection and alignment tools. FBMN enables quantitative analysis and resolution of isomers, including from ion mobility spectrometry.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
The LC–MS2 data for the E. dendroides dataset, along with the MZmine project and parameters used, can be accessed on the MassIVE submission (MSV000080502; Creative Commons CC0 1.0 Universal license). The classical MN and FBMN jobs can be accessed via the GNPS website at https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=189e8bf16af145758b0a900f1c44ff4a and https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=672d0a5372384cff8c47297c2048d789, respectively.
LC–MS2 data for the AGP were downloaded from MassIVE (MSV000080186; Creative Commons CC0 1.0 Universal license) and processed with MZmine (v2.37). The MZmine project along with parameters and export files were deposited (MSV000084095; Creative Commons CC0 1.0 Universal license). The classical MN and FBMN jobs can be accessed at https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=3c27e43d908c4044bace405cc394cd25 and https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=0a8432b5891a48d7ad8459ba4a89969f, respectively.
The LC–MS2 data for the EDTA case are available on the MassIVE submission (MSV00008263; Creative Commons CC0 1.0 Universal license). The classical MN job can be accessed at https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=fbac1a5061ba4ad683a284ef55d45df6. The OpenMS and FBMN jobs are available at https://proteomics2.ucsd.edu/ProteoSAFe/status.jsp?task=83a0a417a49b4b76b61e9a8191a6ea2d at https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=8f40420c11694cf9ab06fdf7a5a4c53b, respectively.
The MS acquisition method, data and parameters used for the processing of the serum analysis with the timsTOF mass spectrometer were deposited (MSV000084402). Classical MN and FBMN jobs can be accessed at https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=f2adc2cf33c646548798d0e285197a96 and https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=0d89db67b0974939a91cb7d5bfe87072, respectively.
Code availability
The FBMN workflow is available as a web interface on the GNPS web platform (https://gnps-quickstart.ucsd.edu/featurebasednetworking/). The workflow code is open source and available on GitHub (https://github.com/CCMS-UCSD/GNPS_Workflows/tree/master/feature-based-molecular-networking/). It is released under the license of The Regents of the University of California San Diego and free for non-profit research (https://github.com/CCMS-UCSD/GNPS_Workflows/blob/master/LICENSE/). The workflow was written in Python (v3.7) and deployed with the ProteoSAFE workflow manager used by GNPS (https://proteomics.ucsd.edu/Software/ProteoSAFe/). We also provide documentation, support, example files and additional information on the GNPS documentation website (https://ccms-ucsd.github.io/GNPSDocumentation/featurebasedmolecularnetworking/). The source code of the GNPSExport module in MZmine is available at https://github.com/mzmine/mzmine2/ under the GNU General Public License. The source code of the GNPSExport tool in OpenMS is available at https://github.com/Bioinformatic-squad-DorresteinLab/OpenMS/under the BSD license. The source code for the GNPSExport custom function for XCMS is available at https://github.com/jorainer/xcms-gnps-tools/ under the GNU General Public License.
References
Watrous, J. et al. Mass spectral molecular networking of living microbial colonies. Proc. Natl Acad. Sci. USA 109, E1743–E1752 (2012).
Quinn, R. A. et al. Molecular networking as a drug discovery, drug metabolism and precision medicine strategy. Trends Pharmacol. Sci. 38, 143–154 (2017).
Traxler, M. F. & Kolter, R. A massively spectacular view of the chemical lives of microbes. Proc. Natl Acad. Sci. USA 109, 10128–10129 (2012).
Wang, M. et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 34, 828–837 (2016).
Frank, A. M. et al. Clustering millions of tandem mass spectra. J. Proteome Res. 7, 113–122 (2008).
Hoffmann, N. et al. mzTab-M: a data standard for sharing quantitative results in mass spectrometry metabolomics. Anal. Chem. 91, 3302–3310 (2019).
Nothias, L.-F. et al. Bioactivity-based molecular networking for the discovery of drug leads in natural product bioassay-guided fractionation. J. Nat. Prod. 81, 758–767 (2018).
Cohen, L. J. et al. Functional metagenomic discovery of bacterial effectors in the human microbiome and isolation of commendamide, a GPCR G2A/132 agonist. Proc. Natl Acad. Sci. USA. 112, E4825–E4834 (2015).
McDonald, D. et al. American Gut: an open platform for citizen-science microbiome research. mSystems 3, e0031–18 (2018).
Röst, H. L. et al. OpenMS: a flexible open-source software platform for mass spectrometry data analysis. Nat. Methods 13, 741–748 (2016).
Pluskal, T., Castillo, S., Villar-Briones, A. & Oresic, M. MZmine 2: modular framework for processing, visualizing and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics 11, 395 (2010).
Bolyen, E. et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857 (2019).
Xia, J., Sinelnikov, I. V., Han, B. & Wishart, D. S. MetaboAnalyst 3.0—making metabolomics more meaningful. Nucleic Acids Res. 43, W251–W257 (2015).
Protsyuk, I., Melnik, A. V., Nothias, L. F. & Rappez, L. 3D molecular cartography using LC–MS facilitated by Optimus and’ili software. Nat. Protoc. 13, 134–154 (2018).
Dührkop, K. et al. SIRIUS 4: a rapid tool for turning tandem mass spectra into metabolite structure information. Nat. Methods 16, 299–302 (2019).
Mohimani, H. et al. Dereplication of peptidic natural products through database search of mass spectra. Nat. Chem. Biol. 13, 30–37 (2017).
van der Hooft, J. J. J., Wandy, J., Barrett, M. P., Burgess, K. E. V. & Rogers, S. Topic modeling for untargeted substructure exploration in metabolomics. Proc. Natl Acad. Sci. USA. 113, 13738–13743 (2016).
Tripathi, A. et al. Chemically-informed analyses of metabolomics mass spectrometry data with qemistree. Preprint at bioRxiv 2020.05.04.077636 (2020) https://doi.org/10.1101/2020.05.04.077636.
Tsugawa, H. et al. A lipidome atlas in MS-DIAL 4. Nat. Biotechnol. https://doi.org/10.1038/s41587-020-0531-2 (2020).
Wang, M. et al. Mass spectrometry searches using MASST. Nat. Biotechnol. 38, 23–26 (2020).
Chambers, M. C. et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 30, 918–920 (2012).
Winnikoff, J. R., Glukhov, E., Watrous, J., Dorrestein, P. C. & Gerwick, W. H. Quantitative molecular networking to profile marine cyanobacterial metabolomes. J. Antibiot. 67, 105–112 (2014).
Olivon, F., Grelier, G., Roussi, F., Litaudon, M. & Touboul, D. MZmine 2 data-preprocessing to enhance molecular networking reliability. Anal. Chem. 89, 7836–7840 (2017).
Ono, K., Demchak, B. & Ideker, T. Cytoscape tools for the web age: D3.js and Cytoscape.js exporters. F1000Res. 3, 143 (2014).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Tsugawa, H. et al. MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 12, 523–526 (2015).
Tautenhahn, R., Böttcher, C. & Neumann, S. Highly sensitive feature detection for high resolution LC/MS. BMC Bioinformatics 9, 504 (2008).
Libiseller, G. et al. IPO: a tool for automated optimization of XCMS parameters. BMC Bioinformatics 16, 118 (2015).
McLean, C. & Kujawinski, E. B. AutoTuner: high fidelity and robust parameter selection for metabolomics data processing. Anal. Chem. 92, 5724–5732 (2020).
Lawson, T. N. et al. msPurity: automated evaluation of precursor ion purity for mass spectrometry-based fragmentation in metabolomics. Anal. Chem. 89, 2432–2439 (2017).
Junker, J. et al. TOPPAS: a graphical workflow editor for the analysis of high-throughput proteomics data. J. Proteome Res. 11, 3914–3920 (2012).
Kuhl, C., Tautenhahn, R., Böttcher, C., Larson, T. R. & Neumann, S. CAMERA: an integrated strategy for compound spectra extraction and annotation of liquid chromatography–mass spectrometry datasets. Anal. Chem. 84, 283–289 (2012).
da Silva, R. R. et al. Propagating annotations of molecular networks using in silico fragmentation. PLoS Comput. Biol. 14, e1006089 (2018).
Ernst, M. et al. MolNetEnhancer: enhanced molecular networks by integrating metabolome mining and annotation tools. Metabolites 9, 144 (2019).
Beauxis, Y. & Genta-Jouve, G. Metwork: a web server for natural products anticipation. Bioinformatics 35, 1795–1796 (2019).
Allen, F., Greiner, R. & Wishart, D. Competitive fragmentation modeling of ESI-MS/MS spectra for putative metabolite identification. Metabolomics 11, 98–110 (2015).
Ruttkies, C., Schymanski, E. L., Wolf, S., Hollender, J. & Neumann, S. MetFrag relaunched: incorporating strategies beyond in silico fragmentation. J. Cheminform. 8, 1–16 (2016).
Ludwig, M. et al. ZODIAC: database-independent molecular formula annotation using Gibbs sampling reveals unknown small molecules. Preprint at bioRxiv https://doi.org/10.1101/842740 (2019).
Dührkop, K. et al. Classes for the masses: systematic classification of unknowns using fragmentation spectra. Preprint at bioRxiv https://doi.org/10.1101/2020.04.17.046672 (2020).
Dührkop, K., Shen, H., Meusel, M., Rousu, J. & Böcker, S. Searching molecular structure databases with tandem mass spectra using CSI:FingerID. Proc. Natl Acad. Sci. USA 112, 12580–12585 (2015).
Gurevich, A. et al. Increased diversity of peptidic natural products revealed by modification-tolerant database search of mass spectra. Nat. Microbiol. 3, 319–327 (2018).
Gerlich, M. & Neumann, S. MetFusion: integration of compound identification strategies. J. Mass Spectrom. 48, 291–298 (2013).
Wandy, J. et al. Ms2lda.org: web-based topic modelling for substructure discovery in mass spectrometry. Bioinformatics 34, 317–318 (2017).
Feunang, Y. D. et al. ClassyFire: automated chemical classification with a comprehensive, computable taxonomy. J. Cheminform. 8, 61 (2016).
Cohen, L. J. et al. Commensal bacteria make GPCR ligands that mimic human signalling molecules. Nature 549, 48–53 (2017).
Simón-Manso, Y. et al. Metabolite profiling of a NIST standard reference material for human plasma (SRM 1950): GC–MS, LC–MS, NMR and clinical laboratory analyses, libraries and web-based resources. Anal. Chem. 85, 11725–11731 (2013).
Meier, F. et al. Online parallel accumulation-serial fragmentation (PASEF) with a novel trapped ion mobility mass spectrometer. Mol. Cell. Proteomics 17, 2534–2545 (2018).
Kind, T. et al. LipidBlast in silico tandem mass spectrometry database for lipid identification. Nat. Methods 10, 755–758 (2013).
Acknowledgements
We gratefully acknowledge financial support from the U.S. National Institutes of Health (NIH) for the Center for Computational Mass Spectrometry grant (P41 GM103484), the reuse of metabolomics data (R03 CA211211) and the tools for rapid and accurate structure elucidation of natural products (R01 GM107550 and U19 AG063744 01) to P.C.D.; the NIH grants R24GM127667 and 1R01LM013115 and a National Science Foundation (NSF) award (ABI 1759980) to N.B.; the European Union’s Horizon 2020 grants 704786 (MSCA-GF to L.-F.N.), 634402 and 777222 (T.A. and I.P.) and a European Research Council Consolidator grant METACELL (T.A.). L.-F.N. was supported by the Center for Microbiome Innovation from the University of California San Diego (support program award). D.P. was supported by the German Research Foundation (DFG; grant no. PE 2600/1). S.N. acknowledges funding from Bundesministerium für Bildung und Forschung (FKZ 031L0107) and the European Commission (EC654241). R.S. acknowledges funding by the German Chemical Industry Fund (FCI) fellowship. H.T. was supported by KAKENHI (18H02432 and 18K19155). A.M.C.-R. was supported by an NSF grant (IOS-1656481) to P.C.D. O.A. acknowledges funding from the Bundesministerium für Ernährung und Landwirtschaft (FKZ 2816501214), the Bundesministerium für Wirtschaft und Energie (FKZ AiF18475N), the Bundesministerium für Bildung und Forschung (FKZ 031A430C) and the European Commission (823839), which also supported F.A. and O.K. S.B. acknowledges funding from Deutsche Forschungsgemeinschaft (BO 1910/20). M.L. was supported by the Deutsche Forschungsgemeinschaft (BO 1910/20-1). J.J.J.v.d.H. was supported by an Accelerating Scientific Discoveries Grant funded by the Netherlands eScience Center (NLeSC; no. ASDI.2017.030). S.N. acknowledges funding from BMBF (grant no. 031L0107) and the European Commission (PhenoMeNal grant EC654241). F.V. was funded by the Department of Navy, Office of Naval Research Multidisciplinary University Research Initiative (MURI) award (N00014-15-1-2809). V.V.P. acknowledges support from the ALSAM Foundation (Therapeutic Innovation Award and L.S. Skaggs Professorship) and the NIH (R35 GM128690). T.P. is a Simons Foundation Fellow of the Helen Hay Whitney Foundation. Z.K. was supported by the project International Mobility of Researchers (CZ.02.2.69/0.0/0.0/16_027/0007990). A.K.J. was supported by the American Society for Mass Spectrometry (Postdoctoral Career Development Award). K.B.K. was supported by a grant from the National Research Foundation (NRF) of Korea (MSIT; NRF-2019R1F1A1058068). H.Y. was supported by the Basic Science Research Program through the NRF grant (NRF- 2018R1C1B6002574). A.L.G. was supported by Vaincre la mucoviscidose and Association Grégory Lemarchal. The work of H.M. was supported by a research fellowship from the Alfred P. Sloan Foundation and an NIH New Innovator Award (DP2GM137413). The authors thank N. Hoffman for maintaining the mzTab-M format. Finally, we acknowledge the continuous feedback from the GNPS community and the contribution of all researchers and associated institutions who are committed to depositing their MS data in public repositories.
Author information
Authors and Affiliations
Contributions
L.-F.N., D.P., M.W. and P.C.D. conceived the method and supervised its implementation and wrote the manuscript. I.P., L.-F.N., M.E. and T.A. created the FBMN prototype in Optimus. M.W., L.-F.N., D.P. and Z.Z. created the FBMN workflow on GNPS. R.S., L.-F.N, M.W., D.P., A.K., M.F., Z.Z., A.S. and T.P. developed the GNPSExport module in MZmine. K.D., A.K., M.L. and S.B. developed the spectral clustering algorithm and SIRIUS export in MZmine. A.S. and L.-F.N. created the GNPSExport tool in OpenMS, with guidance from F.A., O.A. and O.K. J.R. and M.W. created the XCMS export tool. H.T., M.W. and L.-F.N. enabled the integration with MS-DIAL. L.-F.N., A.B., H.N., F.Z. and T.D. enabled the integration with MetaboScape. M.W., G.I., B.S., S.W.M. and J.M. enabled the integration with Progenesis QI. F.V. performed the MS for the plasma and NIST1950SRM samples. A.A.A. performed the MS for the AGP samples. A.K.J., L.-F.N. and A.T. analyzed the results of the plasma samples. J.R. and L.-F.N. performed the XCMS processing of the forensic dataset. L.-F.N. and M.W. created the FBMN documentation. The serum sample analysis in PASEF mode and the data processing with MetaboScape were performed by F.Z., and the subsequent FBMN analysis was performed by L.-F.N. D.P., L.-F.N. and R.d.S. created the MZmine documentation. K.B.K. and H.Y. created the MS-DIAL documentation. F.V., J.M.G., K.W. and A.K.J. prepared the MS-DIAL video tutorial. M.W., R.S. and D.P. prepared the MZmine video tutorials. M.E., R.d.S., J.R., O.M. and S.N. created the XCMS documentation. L.-F.N. and A.S. created the OpenMS documentation. L.-F.N., N.H.N. and T.D. created the MetaboScape documentation. A.M.C.-R. and L.-I.M. documented the FBMN interface workflow. M.N.-E., I.K. and C.M. created the Cytoscape documentation. H.M., A.G., M.W. and L.-F.N. made the integration with DEREPLICATOR. M.W., J.J.J.v.d.H., M.E. and S.R. made the integration with MS2LDA. R.d.S made the integration with NAP. M.M., N.B., X.C., V.V.P., J.P., N.G., R.A.Q., A.A.A., Z.K. and S.N. tested and provided suggestions on how to improve the methods. J.J.J.v.d.H., T.A., A.K.J., T.P., V.V.P., A.L.G., L.-I.M., P.-M.A., S.B. and S.N. improved the manuscript. All authors contributed to the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
P.C.D. is a scientific advisor for Sirenas, Galileio and Cybele and scientific advisor and founder of Ometa labs and Enveda. M.W. is a founder of Ometa Labs. T.P. is a consultant for Ginkgo Bioworks. A.A.A. is a consultant for Ometa Labs. T.A. is on the Scientific Advisory Board of SCiLS, a Bruker company. K.D., M.L., M.F. and S.B. are founders of Bright Giant. A.B., S.W.M., H.N. and F.Z. are employees of Bruker Daltonics. G.I., J.M. and B.S. are employees of Waters.
Additional information
Peer review information Arunima Singh was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–19, Supplementary Notes 1–4 and Supplementary Tables 1 and 2.
Rights and permissions
About this article
Cite this article
Nothias, LF., Petras, D., Schmid, R. et al. Feature-based molecular networking in the GNPS analysis environment. Nat Methods 17, 905–908 (2020). https://doi.org/10.1038/s41592-020-0933-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-020-0933-6
This article is cited by
-
Apiospora arundinis, a panoply of carbohydrate-active enzymes and secondary metabolites
IMA Fungus (2024)
-
Genome sequencing and molecular networking analysis of the wild fungus Anthostomella pinea reveal its ability to produce a diverse range of secondary metabolites
Fungal Biology and Biotechnology (2024)
-
Murraya koenigii (L.) Sprengel seeds and pericarps in relation to their chemical profiles: new approach for multidrug resistant Acinetobacter baumannii ventilator-associated pneumonia
Applied Biological Chemistry (2024)
-
A multiplex metabolomic approach for quality control of Spirulina supplement and its allied microalgae (Amphora & Chlorella) assisted by chemometrics and molecular networking
Scientific Reports (2024)
-
Bile salt hydrolase acyltransferase activity expands bile acid diversity
Nature (2024)