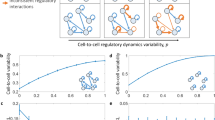
Abstract
Single-cell genomics has transformed our ability to examine cell fate choice. Examining cells along a computationally ordered ‘pseudotime’ offers the potential to unpick subtle changes in variability and covariation among key genes. We describe an approach, scHOT—single-cell higher-order testing—which provides a flexible and statistically robust framework for identifying changes in higher-order interactions among genes. scHOT can be applied for cells along a continuous trajectory or across space and accommodates various higher-order measurements including variability or correlation. We demonstrate the use of scHOT by studying coordinated changes in higher-order interactions during embryonic development of the mouse liver. Additionally, scHOT identifies subtle changes in gene–gene correlations across space using spatially resolved transcriptomics data from the mouse olfactory bulb. scHOT meaningfully adds to first-order differential expression testing and provides a framework for interrogating higher-order interactions using single-cell data.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All data analysis was performed on publicly available data. The liver developmental dataset is described at https://sydneybiox.github.io/scMerge/articles/case_study/Mouse_Liver_Data.html and instructions to download the data are available in the analysis scripts at https://github.com/MarioniLab/scHOT2019.
The MOB data was downloaded from the Spatial Research website https://www.spatialresearch.org/resources-published-datasets/doi-10-1126science-aaf2403/ and count matrix data and H&E stained brightfield image related to MOB replicate 11 was downloaded. Source data are provided with this paper.
Code availability
Instructions to access data, software, and scripts to perform the analysis presented here is available at https://github.com/MarioniLab/scHOT2019. scHOT is available as a Bioconductor R package https://bioconductor.org/packages/scHOT with detailed vignette available.
References
Pijuan-Sala, B. et al. A single-cell molecular map of mouse gastrulation and early organogenesis. Nature 566, 490–495 (2019).
Cao, J. et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502 (2019).
Briggs, J. A. et al. The dynamics of gene expression in vertebrate embryogenesis at single-cell resolution. Science 360, 1–16 (2018).
Wagner, D. E. et al. Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science 360, 981–987 (2018).
Polioudakis, D. et al. A single-cell transcriptomic atlas of human neocortical development during mid-gestation. Neuron 103, 785–801 (2019).
Mohammed, H. et al. Single-cell landscape of transcriptional heterogeneity and cell fate decisions during mouse early gastrulation. Cell Rep. 20, 1215–1228 (2017).
Mojtahedi, M. et al. Cell fate decision as high-dimensional critical state transition. PLoS Biol. 14, e2000640 (2016).
Basu, S., Kumbier, K., Brown, J. B. & Yu, B. Iterative random forests to discover predictive and stable high-order interactions. Proc. Natl Acad. Sci. USA 115, 1943–1948 (2018).
Bageritz, J. et al. Gene expression atlas of a developing tissue by single cell expression correlation analysis. Nat. Methods 16, 750–756 (2019).
Ståhl, P. L. et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353, 78–82 (2016).
Lubeck, E., Coskun, A. F., Zhiyentayev, T., Ahmad, M. & Cai, L. Single-cell in situ RNA profiling by sequential hybridization. Nat. Methods 11, 360–361 (2014).
Chen, K. H., Boettiger, A. N., Moffitt, J. R., Wang, S. & Zhuang, X. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090 (2015).
Street, K. et al. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics 19, 477 (2018).
Campbell, K. R. & Yau, C. A descriptive marker gene approach to single-cell pseudotime inference. Bioinformatics 35, 28–35 (2018).
Haghverdi, L., Büttner, M., Wolf, F. A., Buettner, F. & Theis, F. J. Diffusion pseudotime robustly reconstructs lineage branching. Nat. Methods 13, 845–848 (2016).
Trapnell, C. et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014).
Van den Berge, K. et al. Trajectory-based differential expression analysis for single-cell sequencing data. Nat. Commun. 11, 1201 (2020).
Lönnberg, T. et al. Single-cell RNA-seq and computational analysis using temporal mixture modelling resolves Th1/Tfh fate bifurcation in malaria. Sci. Immunol. 2, eaal2192 (2017).
Qiu, X. et al. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979–982 (2017).
Fuller, T. F. et al. Weighted gene coexpression network analysis strategies applied to mouse weight. Mamm. Genome 18, 463–472 (2007).
Treutlein, B. et al. Dissecting direct reprogramming from fibroblast to neuron using single-cell RNA-seq. Nature 534, 391–395 (2016).
Chen, S. & Mar, J. C. Evaluating methods of inferring gene regulatory networks highlights their lack of performance for single cell gene expression data. BMC Bioinf. 19, 232 (2018).
Ghazanfar, S., Strbenac, D., Ormerod, J. T., Yang, J. Y. H. H. & Patrick, E. DCARS: differential correlation across ranked samples. Bioinformatics 35, 823–829 (2019).
Svensson, V., Teichmann, S. A. & Stegle, O. SpatialDE: identification of spatially variable genes. Nat. Methods 15, 343–346 (2018).
Nitzan, M., Karaiskos, N., Friedman, N. & Rajewsky, N. Gene expression cartography. Nature 576, 132–137 (2019).
Yang, L. et al. A single-cell transcriptomic analysis reveals precise pathways and regulatory mechanisms underlying hepatoblast differentiation. Hepatology 66, 1387–1401 (2017).
Dong, J. et al. Single-cell RNA-seq analysis unveils a prevalent epithelial/mesenchymal hybrid state during mouse organogenesis. Genome Biol. 19, 31 (2018).
Su, X. et al. Single-cell RNA-Seq analysis reveals dynamic trajectories during mouse liver development. BMC Genomics 18, 946 (2017).
Camp, J. G. et al. Multilineage communication regulates human liver bud development from pluripotency. Nature 546, 533–538 (2017).
Oikawa, T. et al. Sall4 regulates cell fate decision in fetal hepatic stem/progenitor cells. Gastroenterology 136, 1000–1011 (2009).
Tanaka, M. et al. Mouse hepatoblasts at distinct developmental stages are characterized by expression of EpCAM and DLK1: drastic change of EpCAM expression during liver development. Mech. Dev. 126, 665–676 (2009).
Sugimoto, N. et al. Identification of novel human Cdt1-binding proteins by a proteomics approach: proteolytic regulation by APC/CCdh1. Mol. Biol. Cell 19, 1007–1021 (2008).
Zape, J. P., Lizama, C. O., Cautivo, K. M. & Zovein, A. C. Cell cycle dynamics and complement expression distinguishes mature haematopoietic subsets arising from hemogenic endothelium. Cell Cycle 16, 1835–1847 (2017).
Thakurela, S. et al. Gene regulation and priming by topoisomerase IIα in embryonic stem cells. Nat. Commun. 4, 2478 (2013).
Rialland, M., Sola, F. & Santocanale, C. Essential role of human CDT1 in DNA replication and chromatin licensing. J. Cell Sci. 115, 1435–1440 (2002).
Lin, D. M. et al. Spatial patterns of gene expression in the olfactory bulb. Proc. Natl Acad. Sci. USA 101, 12718–12723 (2004).
Fan, X. L., Zhang, J. S., Zhang, X. Q., Yue, W. & Ma, L. Differential regulation of β-arrestin 1 and β-arrestin 2 gene expression in rat brain by morphine. Neuroscience 117, 383–389 (2003).
Macias, M. et al. Spatiotemporal characterization of mTOR kinase activity following kainic acid induced status epilepticus and analysis of rat brain response to chronic rapamycin treatment. PLoS ONE 8, e64455 (2013).
Wilson, P. O. et al. The immunolocalization of protein gene product 9.5 using rabbit polyclonal and mouse monoclonal antibodies. Br. J. Exp. Pathol. 69, 91–104 (1988).
Gray, N. W. et al. Dynamin 3 is a component of the postsynapse, where it interacts with mGluR5 and homer. Curr. Biol. 13, 510–515 (2003).
Kendall, R. T. et al. Arrestin-dependent angiotensin AT 1 receptor signaling regulates akt and mTor-mediated protein synthesis. J. Biol. Chem. 289, 26155–26166 (2014).
Girnita, L. et al. β-Arrestin is crucial for ubiquitination and down-regulation of the insulin-like growth factor-1 receptor by acting as adaptor for the MDM2 E3 ligase. J. Biol. Chem. 280, 24412–24419 (2005).
Bhatnagar, A. et al. The dynamin-dependent, arrestin-independent internalization of 5-hydroxytryptamine 2A (5-HT 2A) serotonin receptors reveals differential sorting of arrestins and 5-HT 2A receptors during endocytosis. J. Biol. Chem. 276, 8269–8277 (2001).
Jacque, C. M., Collet, A., Raoul, M., Monge, M. & Gumpel, M. Functional maturation of the oligodendrocytes and myelin basic protein expression in the olfactory bulb of the mouse. Dev. Brain Res. 21, 277–282 (1985).
Guillemin, A., Duchesne, R., Crauste, F., Gonin-Giraud, S. & Gandrillon, O. Drugs modulating stochastic gene expression affect the erythroid differentiation process. PLoS ONE 14, e0225166 (2019).
Moris, N. et al. Histone acetyltransferase KAT2A stabilizes pluripotency with control of transcriptional heterogeneity. Stem Cells 36, 1828–1838 (2018).
Richard, A. et al. Single-cell-based analysis highlights a surge in cell-to-cell molecular variability preceding irreversible commitment in a differentiation process. PLoS Biol. 14, e1002585 (2016).
Semrau, S. et al. Dynamics of lineage commitment revealed by single-cell transcriptomics of differentiating embryonic stem cells. Nat. Commun. 8, 1096 (2017).
Stumpf, P. S. et al. Stem cell differentiation as a non-Markov stochastic process. Cell Syst. 5, 268–282.e7 (2017).
Wiesner, K., Teles, J., Hartnor, M. & Peterson, C. Haematopoietic stem cells: entropic landscapes of differentiation. Interface Focus 8, 20180040 (2018).
Lin, Y. et al. scMerge leverages factor analysis, stable expression, and pseudoreplication to merge multiple single-cell RNA-seq datasets. Proc. Natl Acad. Sci. USA 116, 9775–9784 (2019).
Lin, Y. et al. Evaluating stably expressed genes in single cells. Gigascience 8, 229815 (2019).
Brennecke, P. et al. Accounting for technical noise in single-cell RNA-seq experiments. Nat. Methods 10, 1093–1095 (2013).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B. 57, 289–300 (1995).
Langfelder, P., Zhang, B. & Horvath, S. Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics 24, 719–720 (2008).
Ashburner, M. et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29 (2000).
The Gene Ontology Consortium. The gene ontology resource: 20 years and still GOing strong. Nucleic Acids Res. 47, D330–D338 (2019).
Székely, G. J., Rizzo, M. L. & Bakirov, N. K. Measuring and testing dependence by correlation of distances. Ann. Stat. 35, 2769–2794 (2007).
Ghazanfar, S. & Yang, J. Y. H. Characterizing mutation–expression network relationships in multiple cancers. Comput. Biol. Chem. 63, 73–82 (2016).
Pimentel, R. S., Niewiadomska-Bugaj, M. & Wang, J. C. Association of zero-inflated continuous variables. Stat. Probab. Lett. 96, 61–67 (2015).
Acknowledgements
We thank all our colleagues, particularly at Cancer Research UK Cambridge Institute and The University of Sydney School of Mathematics and Statistics for their support and intellectual engagement. In particular, we acknowledge P. Yang and M. Morgan for their helpful discussion. We acknowledge the following sources of funding: Royal Society Newton International Fellowship (NIF\R1\181950) and funding from the Judith and Coffey Life Laboratory at the Charles Perkins Centre to S.G.; Australia NHMRC Career Developmental Fellowship (APP1111338) to J.Y.H.Y.; Research Training Program Tuition Fee Offset and Stipend Scholarship and Chen Family Research Scholarship to Y.L.; Australian Research Council Discovery Early Career Researcher Award (DE200100944) funded by the Australian Government to E.P.; NIH grant (R21DC015107) to D.M.L.; SJTU-USYD Translate Medicine Fund-Systems Biomedicine (AF6260003) to J.Y.H.Y., X.S. and Z.G.H.; National Natural Science Foundation of China (81802806) to X.S.; and Core funding from EMBL and Cancer Research UK (award no. 17197) to J.C.M. The funding source had no role in the study design; in the collection, analysis and interpretation of data, in the writing of the manuscript and in the decision to submit the manuscript for publication.
Author information
Authors and Affiliations
Contributions
S.G. conceived the study with input from E.P., J.Y.H.Y. and J.C.M. S.G. developed the method and software and performed data analysis with input from Y.L. S.G., D.M.L. and X.S. interpreted the results with input from Z.G.H. and J.C.M. S.G., J.C.M. and J.Y.H.Y. wrote the manuscript. All authors read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Lin Tang was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Variability testing along hepatoblast branch using scHOT.
a, Scatter and ribbon plot of all significant genes showing loss of variability along hepatoblast branch (176 cells), ribbon width corresponding to adding and subtracting the weighted standard deviation from the weighted mean. b, Scatter and ribbon plot of all significant genes showing gain of variability along hepatoblast branch, with ribbon width as in Panel A.
Extended Data Fig. 2 Correlation testing of cholangiocyte branch using scHOT.
a, Line plots of clustered significant scHOT gene-pairs with FDR adjusted P-value < 0.2 for full cholangiocyte branch (308 cells). Vertical dashed line indicates trajectory branchpoint. b, Gene ontology functional enrichment using one-sided Fisher’s Exact Test barplots for all scHOT cholangiocyte clusters, grey bar color corresponds to FDR adjusted P-value < 0.05. Gene sample sizes indicated in each plot title. c, Gene ontology functional enrichment using one-sided Fisher’s Exact Test barplots for all scHOT hepatocyte clusters, with color as in panel B. Gene sample sizes indicated in each plot title.
Extended Data Fig. 3 Correlation testing of spatial data using scHOT.
a, Spatial expression plots (262 spatially resolved positions) of two gene-pairs Arrb1 and Uchl1 as well as Arrb1 and Dnm3 which are not significantly differentially expressed across space using scHOT, but are significantly differentially correlated across space using scHOT with FDR adjusted P-value < 0.2. The third plot shows the local spatial correlation estimated for these two genes, recapitulating the layered pattern of the olfactory bulb. b, Spatial maps of mean local correlation and Gene Ontology functional enrichment using one-sided Fisher’s Exact Test barplots for all MOB scHOT clusters, gray bar color corresponds to FDR adjusted P-value < 0.05. Gene sample sizes indicated in each plot title.
Extended Data Fig. 4 Highly variable gene selection and permutation testing for scHOT.
a, HVG selection for Developmental Liver Data. b, HVG selection for Spatial Transcriptomics analysis. c, Global correlation and null scHOT correlation test statistics for sampled gene pairs in both hepatocyte and cholangiocyte branches (268 and 265 gene pairs respectively). d, Global correlation and null scHOT correlation test statistics for sampled gene pairs (172 gene pairs) for spatial MOB data. e, Scatterplot of -log10(P-values) for differential correlation testing (22,155 gene pairs) of the liver hepatocyte branch, calculated using 10,000 permutations for each gene-pair (x-axis), and estimated using borrowed permutations over a subset of gene-pairs (y-axis). Black solid line corresponds to y = x, grey dashed lines correspond to unadjusted P-values of 0.05, and the solid red curve corresponds to the fitted loess curve.
Extended Data Fig. 5 Further scHOT method description.
a, Illustrative example showing testing for correlation differences in three distinct groups. A set of local higher order statistics are calculated, and significance is compared by repeatedly permuting samples (grey boxplots). Illustrative example shows the set of local estimates of higher order statistics are combined using the sample standard deviation to assess how variable they are between groups. b, Possible schemes for the testing scaffold using gene networks, including: i) a gene-gene network; ii) a gene set scaffold where all pairwise combinations within a gene set are included; and iii) selected genes of interest versus all others. c, Examples of weighted higher order functions including weighted Pearson correlation, weighted Spearman correlation, weighted variance. Note that any user defined function can be used.
Extended Data Fig. 6 scHOT stability under parameter choices.
a, Spearman correlation map of -log10(P-values) of scHOT differential correlation testing (22,155 gene pairs) in hepatocyte branch with different choices of triangular span, from 0.05 to 0.70 in steps of 0.05. b, UpSet plot of 509 significant gene-pairs (FDR adjusted P-value < 0.2) from each scHOT testing scheme as in panel a. c, Spearman correlation map of -log10(P-values) of scHOT differential correlation testing ( 903 gene pairs) in MOB with different choice of spatial span. d, UpSet plot of 181 significant gene-pairs from each testing scheme as in panel c. e, Density plot of slopes of local correlation patterns of gene-pairs selected with low span (0.35 and below, colored blue) and high span (0.35 and above, colored red), dotted line shows the density over all slopes. f, Spearman correlation map of -log10(P-values) of scHOT differential correlation testing (22,155 gene pairs) in hepatocyte branch with different choices of higher order statistic, MIC—maximal information criterion, BDC—Brownian distance correlation, Pearson_block and Spearman_block refer to Pearson and Spearman correlation respectively, applied in a block-weighted context. g, UpSet plot of 367 significant gene pairs from each testing scheme as in panel f.
Extended Data Fig. 7 Robustness of scHOT results.
a, Scatterplots of -log10(P-values) for hepatoblast to hepatocyte correlation scHOT testing (22,155 gene pairs) against the inclusion frequency of gene-pairs with FDR adjusted P-values < 0.2 for repeated subsampling without replacement of 90%, 80%, 70%, 60%, and 50% of the cells from the trajectory. Red points correspond to those selected as FDR adjusted P-value < 0.2 criteria for the full dataset. b, Spearman correlation map of -log10(P-values) of each subsampling strategy. c, Quantile-quantile line plots of -log10(P-values) for the full data (x-axis) and -log10(P-values) for each subsampling scenario (y-axis), split by subsampling percentage. Red lines correspond to y = x.
Supplementary information
41592_2020_885_MOESM2_ESM.xls
Supplementary Table 1 a, Significantly differentially variable genes using scHOT for the hepatoblast to branchpoint trajectory analysis (176 cells), with columns for the gene, the observed scHOT test statistic, unadjusted P value, FDR adjusted P value, gain or loss of variability and local variability and the start and end of the trajectory. b, Significantly differentially correlated gene pairs using scHOT for the hepatoblast to hepatocyte trajectory analysis (408 cells), with columns for the gene pair, the observed scHOT test statistic, the global Spearman correlation value, unadjusted P value, number of permutations used for P value estimation, minimum and maximum values of global correlation for which P value estimation was used, FDR adjusted P value, cluster number and local correlation at the start, branchpoint and end of the trajectory.
Source data
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Rights and permissions
About this article
Cite this article
Ghazanfar, S., Lin, Y., Su, X. et al. Investigating higher-order interactions in single-cell data with scHOT. Nat Methods 17, 799–806 (2020). https://doi.org/10.1038/s41592-020-0885-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-020-0885-x
This article is cited by
-
DELVE: feature selection for preserving biological trajectories in single-cell data
Nature Communications (2024)
-
SRTsim: spatial pattern preserving simulations for spatially resolved transcriptomics
Genome Biology (2023)
-
Decoding the tumor microenvironment with spatial technologies
Nature Immunology (2023)
-
SpatialDM for rapid identification of spatially co-expressed ligand–receptor and revealing cell–cell communication patterns
Nature Communications (2023)
-
Principles and challenges of modeling temporal and spatial omics data
Nature Methods (2023)