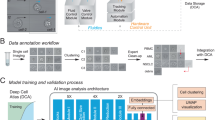
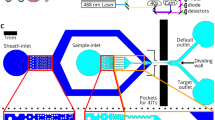
Abstract
Although label-free cell sorting is desirable for providing pristine cells for further analysis or use, current approaches lack molecular specificity and speed. Here, we combine real-time fluorescence and deformability cytometry with sorting based on standing surface acoustic waves and transfer molecular specificity to image-based sorting using an efficient deep neural network. In addition to general performance, we demonstrate the utility of this method by sorting neutrophils from whole blood without labels.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The data supporting findings of this study are available on figshare (https://doi.org/10.6084/m9.figshare.11302595). Source data for Figs. 1–3 and Extended Data Figs. 2–8 are provided with the paper.
Code availability
Shape-Out and AID are open source and available on GitHub at URLs provided in the Methods. Shape-In is commercially available from Zellmechanik Dresden GmbH. The C++ code used for sorting is custom-built for the hardware used in the set-up and available onsite to interested parties.
References
Dainiak, M. B., Kumar, A., Galaev, I. Y. & Mattiasson, B. in Cell Separation 1–18 (Springer, 2007).
Wyatt Shields, Iv,C., Reyes, C. D. & López, G. P. Microfluidic cell sorting: a review of the advances in the separation of cells from debulking to rare cell isolation. Lab Chip 15, 1230–1249 (2015).
Baron, C. S. et al. Cell type purification by single-cell transcriptome-trained sorting. Cell 179, 527–542 (2019).
Stamm, C. et al. Autologous bone-marrow stem-cell transplantation for myocardial regeneration. Lancet 361, 45–46 (2003).
Bartsch, U. et al. Retinal cells integrate into the outer nuclear layer and differentiate into mature photoreceptors after subretinal transplantation into adult mice. Exp. Eye Res. 86, 691–700 (2008).
Miltenyi, S., Müller, W., Weichel, W. & Radbruch, A. High gradient magnetic cell separation with MACS. Cytometry 11, 231–238 (1990).
Bonner, W. A., Hulett, H. R., Sweet, R. G. & Herzenberg, L. A. Fluorescence activated cell sorting. Rev. Sci. Instrum. 43, 404–409 (1972).
Shapiro, H. M. Practical Flow Cytometry (John Wiley & Sons, 2003).
Preira, P. et al. Passive circulating cell sorting by deformability using a microfluidic gradual filter. Lab Chip 13, 161–170 (2013).
Wang, G. et al. Microfluidic cellular enrichment and separation through differences in viscoelastic deformation. Lab Chip 15, 532–540 (2015).
Beech, J. P., Holm, S. H., Adolfsson, K. & Tegenfeldt, J. O. Sorting cells by size, shape and deformability. Lab Chip 12, 1048–1051 (2012).
Otto, O. et al. Real-time deformability cytometry: on-the-fly cell mechanical phenotyping. Nat. Methods 12, 199–202 (2015).
Toepfner, N. et al. Detection of human disease conditions by single-cell morpho-rheological phenotyping of blood. eLife 7, e29213 (2018).
Nitta, N. et al. Intelligent image-activated cell sorting. Cell 175, 266–276 (2018).
Rosendahl, P. et al. Real-time fluorescence and deformability cytometry. Nat. Methods 15, 355 (2018).
Nawaz, A. A. et al. Acoustofluidic fluorescence activated cell sorter. Anal. Chem. 87, 12051–12058 (2015).
Girault, M. et al. An on-chip imaging droplet-sorting system: a real-time shape recognition method to screen target cells in droplets with single cell resolution. Sci. Rep. 7, 40072 (2017).
Girardo, S. et al. Standardized microgel beads as elastic cell mechanical probes. J. Mater. Chem. B 6, 6245–6261 (2018).
Mietke, A. et al. Extracting cell stiffness from real-time deformability cytometry: theory and experiment. Biophys. J. 109, 2023–2036 (2015).
Mokbel, M. et al. Numerical simulation of real-time deformability cytometry to extract cell mechanical properties. ACS Biomater. Sci. Eng. 3, 2913–2962 (2017).
Hartono, D. et al. On-chip measurements of cell compressibility via acoustic radiation. Lab Chip 11, 4072–4080 (2011).
Gustafson, M. P. et al. A method for identification and analysis of non-overlapping myeloid immunophenotypes in humans. PLoS One 10, e0121546 (2015).
Bashant, K. R. et al. Real-time deformability cytometry reveals sequential contraction and expansion during neutrophil priming. J. Leukoc. Biol. 105, 1143–1153 (2019).
Di Carlo, D., Irimia, D., Tompkins, R. G. & Toner, M. Continuous inertial focusing, ordering, and separation of particles in microchannels. Proc. Natl Acad. Sci. USA 104, 18892–18897 (2007).
Ding, X. et al. Surface acoustic wave microfluidics. Lab Chip 13, 3626–3649 (2013).
Bradski, G. The OpenCV library. Dr Dobb’s J. Softw. Tools 25, 120–126 (2000).
Herold, C. Mapping of deformation to apparent Young’s modulus in real-time deformability cytometry. Preprint at https://arxiv.org/abs/1704.00572 (2017).
Kräter, M. et al. AIDeveloper: deep learning image classification in life science and beyond. Preprint at https://doi.org/10.1101/2020.03.03.975250 (2020).
Nickolls, J., Buck, I., Garland, M. & Skadron, K. Scalable parallel programming with CUDA. Queue 6, 40 (2008).
Abadi, M. et al. TensorFlow: large-scale machine learning on heterogeneous dstributed systems. Preprint at https://arxiv.org/abs/1603.04467 (2016).
Al-Rfou, R. et al. Theano: a Python framework for fast computation of mathematical expressions. Preprint at https://arxiv.org/abs/1605.02688 (2016).
Glaubitz, M. et al. A novel contact model for AFM indentation experiments on soft spherical cell-like particles. Soft Matter 10, 6732–6741 (2014).
Acknowledgements
We thank D. Soteriou (MPL Erlangen) for engaging discussions, and C. Schweitzer (MPL Erlangen) and I. Richter (TU Dresden) for technical assistance. We further thank the Microstructure Facility at the Center for Molecular and Cellular Bioengineering (CMCB) at Technische Universität Dresden (in part funded by the State of Saxony and the European Fund for Regional Development) for the production of the microgel beads and hosting the chip fabrication. The HL60/S4 cells were a kind gift of D. E. Olins and A. L. Olins (University of New England), and Kc167 cells were a kind gift of B. Baum (University College London). The authors acknowledge funding from the Alexander von Humboldt-Stiftung (Alexander von Humboldt Professorship to J.G.), ERC Starting Grant (starting grant LightTouch no. 282060 to J.G.), European Commission through LAPASO ITN project (EU FP7/2007-2013, no. 607350), a DKMS ‘Mechthild Harf Research Grant’ (DKMS-SLS-MHG-2016-02 to A.J.) and German Research Foundation (no. GU 612/5-1 to J.G.).
Author information
Authors and Affiliations
Contributions
J.G. conceived the project. A.A.N. developed and built the microfluidic chips and SSAW actuators. M.N., P.R. and A.A.N. integrated SSAW sorting into the experimental set-up. M.N., with the contributions of P.R. and C.H., developed the software for real-time sorting. S.A. performed flow simulations validating chip design. A.A.N., with the contributions of M.H., M. Kräter, A.J., P.R., M.U., N.T. and M. Kubankova, performed the sorting experiments with beads, cells and blood samples. M. Kräter., A.J., M.H. and N.T. prepared the blood samples and developed cell staining protocols. M.H. developed the DNN analysis and performed the artificial-intelligence sorting experiments. S.G. and R.G. produced the polyacrylamide beads. S.G., R.G. and F.R. provided advice and technical support for chip production. A.T. performed AFM measurements. M.U., M.H. and A.A.N. analyzed the data. P.M. developed Shape-Out analysis software. M.U. visualized the data and prepared figures. M.U., with the support of J.G., P.R., M.H. and A.A.N. prepared the initial version of the manuscript. All authors revised and edited the manuscript. J.G. and A.J. acquired funding.
Corresponding authors
Ethics declarations
Competing interests
M.U., M. Kräter, N.T., M. Kubankova, R.G., S.A., F.R., A.T., S.G. and A.J. declare no competing interests. A.A.N., M.H., M.N. and J.G. have applied for patent protection of this method. P.R., C.H. and P.M. work for Zellmechanik Dresden GmbH, the company which sells devices based on RT-FDC technology. P.R. and C.H. hold shares of Zellmechanik Dresden GmbH.
Additional information
Peer review information Rita Strack was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Extended Data Fig. 1 Visualization of bead trajectories in the soRT-FDC chip in the presence and absence of SSAW actuation.
a,b, An image of soRT-FDC chip showing trajectory taken by beads when SSAW actuation is off (a) and when SSAW actuation is on (b). The images were created by a minimum intensity projection of 700 frames taken over 350 ms. The flow direction is from left to right. The red line indicates the center of the channel. Note the slight off-center position of the bifurcation point.
Extended Data Fig. 2 Determination of bead sizes using RT-FDC, soRT-FDC, and flow cytometry (FC).
A mixture of beads of four different sizes, each made of different material, was analyzed with (so)RT-FDC and FC (BD LSR II, BD Biosciences). We gated for the different bead populations using two-dimensional analysis combining size and brightness in RT-FDC (a) and soRT-FDC (b), and forward scatter area (FSC-A) and side scatter area (SSC-A) in FC (c). In a–c color map in scatter plots represents event density, the histograms of area and FSC-A are shown on top of scatter plots, and the histograms of brightness and SSC-A on the right-hand side of the scatter plots. Color-coded solid lines represent Gaussian fits to the gated populations of beads. For illustration purposes, data in a–c were resampled to equalize the relative content of each bead population. (d) A summary of bead properties, including bead material, and bead sizes as specified by the supplier, measured with RT-FDC (using 40 × objective and glass substrate) and soRT-FDC (using 20× objective, lithium niobate substrate and an extra polarizer), as well as measured with FC. The number of events, n, analyzed for each bead type using respective methods is indicated in the table. Note that the bead size determined by FC is a dimensionless value and not a physical dimension. Furthermore, the relative bead sizes determined with FC do not follow the size order specified by the supplier, as indicated by increasing grey levels in the background. The experiments in a–c were repeated independently 2, 3, and 1 times, respectively, with similar results.
Extended Data Fig. 3 Further examples of bead sorting with two-dimensional gates performed using soRT-FDC.
a, Sorting for deformation together with maximum fluorescence intensity in a mixture of fluorescent and non-fluorescent polyacrylamide beads with different mechanical properties. b, Sorting for brightness and size in a mixture of beads with different diameters, each made of different material (see Extended Data Fig 2c, d). The color map in scatter plots represents event density. The histograms of features presented in the scatter plots are shown on top and on the right of the corresponding scatter plots, the histograms were fit with superpositions of Gaussian functions (solid lines). The gates used for sorting are outlined in green. Percentages on scatter plots indicate the fraction of beads in the sorting gate. The experiments in a and b were repeated independently 1 and 3 times, respectively, with similar results.
Extended Data Fig. 4 Size-based cell sorting from a mixture of two cell lines.
a, Deformation-size scatter plot of a 1:4 mixture of Kc167 (smaller size) and HL60/S4 (bigger size) cells in the initial mixture. b, Deformation-size scatter plot of sample collected in the target. Kc167 cells were enriched 4-fold to 88.2%. The green shaded rectangle in both plots indicates the sorting gate (size 25–77 µm2, deformation 0–0.15). The color map in scatter plots represents event density. The histograms of cell size are shown on the right of the corresponding scatter plots, the histograms were fit with superpositions of Gaussian functions (solid lines). The gates used for sorting are outlined in green. Percentages on scatter plots indicate the fraction of cells in the sorting gate. Similar experiment, with gating for either HL60/S4 cells or Kc167 cells, was performed independently 4 times with comparable results.
Extended Data Fig. 5 Deformation-based sorting of RBCs.
a, Deformation-size scatter plot for initial measurement of whole diluted blood. The colored rectangles indicate the gates used for RBCs with high (green gate) and low (magenta gate) deformation during two subsequent sorting experiments. (b, c) Deformation-size scatter plot of sample collected in the target when sorting for cells with low deformation (b), 3.0-fold enrichment and 91.3% purity was observed in this case, and when sorting for cells with low deformation (c), 2.8-fold enrichment and 50.3% purity was obtained. The color map in scatter plots represents event density. The histograms of deformation are shown on the right of the corresponding scatter plots, the histograms were fit with Gaussian functions (solid lines). The gates used for sorting in b and c are outlined in green and magenta, respectively. Percentages on scatter plots indicate the fraction of cells in the sorting gate. This experiment was repeated independently 3 times with similar results.
Extended Data Fig. 6 Proliferation and viability of HL60/S4 cells after exposure to SSAW in a soRT-FDC experiment.
a,b, Count of cells prepared as for soRT-FDC experiment but not loaded onto the chip (initial), cells run through soRT-FDC chip but not exposed to SSAW (default), as well as cells exposed to SSAW that were collected in the target outlet (target) cultured over 4 days expressed as absolute cell count (a), and relative count with respect to the number of seeded cells (b). The number of seeded cells was equal to 70,400 for target, and 100,000 for both initial and default samples. (c) Viability of cells in cultures described above evaluated by trypan blue staining. Similar experiment was performed independently 2 times with comparable results.
Extended Data Fig. 7 AFM indentation measurement of HL60/S4 cell stiffness after exposure to SSAW in a soRT-FDC experiment.
Box plots of the apparent Young’s modulus measured for initial, default, and target samples by AFM. The initial sample was prepared as for the soRT-FDC experiment but not loaded onto the chip, the default sample was collected in the default outlet, that is, run through the sorting chip but not exposed to SSAW, and the target sample corresponds to cells exposed to SSAW. The number of cells analyzed for each condition, n, is indicated in the plot. Boxes extend from 25th to 75th percentiles, with a line at the median. Whiskers span 10th to 90th percentile. Scattered data points correspond to outliers. Datasets were compared using a Kruskal-Wallis test with a Dunn’s multiple comparisons test. The obtained p-values are reported on the graph.
Extended Data Fig. 8 Verification of brightness-based sorting of neutrophils from RBC-depleted blood with CD66 and CD14 surface markers.
a, CD66 and CD14 surface marker expression for initial (left) and target (right) samples measured with RT-FDC. b, Brightness-cell size scatter plots as in Fig. 2g, h with CD66+/CD14− cells (putative neutrophils) indicated in orange. Sorting gate is outlined with dashed green line. This experiment was performed independently 3 times with similar results.
Supplementary Information
Supplementary Information
Supplementary Tables 1–7, Supplementary Notes 1–5 and Supplementary references
Supplementary Video 1
Size-based sorting of a mixture of Kc167 and HL60/S4 cell lines using soRT-FDC. The video presents a screen capture of the sorting software window taken during the experiment. The gate of cross-sectional area 25–77 µm2 was set to select for the smaller cells (Kc167) (Extended Data Fig. 4). The top panel presents an image of the sorting chip. The scale marks in the lower right are spaced 20 µm apart. The analysis ROI is marked with a white rectangle. The SSAW sorting region as well as the target and default outlets are indicated with text labels. Before the sorting starts at around 4 s, all cells are directed into the default outlet. Upon sorting start, some of the cells are deflected towards the target outlet. Below the chip image, the bright-field images of cells taken in the ROI are shown only for cells classified for sorting. Next to it, the thresholded and binarized images of all detected cells are displayed. The control panel on the lower left shows several real-time metrics, including total number of sorted cells (Cells sorted), total number of detected cells (Cells detected), the number of detected cells per seconds (Cells detected [1/s]) and the number of sorted cells per second (Cells sorted [1/s]). The real-time scatter plot of cell size (area) versus. deformation of all detected cells is shown on the right-hand side and restarted after ca. 500 events. This screen capture was recorded with VLC media player at approximately 25 frames per s, 100 times lower than the frame rate used for analyzing the cells. A similar experiment, with gating either for HL60/S4 or Kc167 cells, was performed independently four times with similar results, and the screen capture was performed once.
Supplementary Video 2
Brightness-based sorting of neutrophils from RBC-depleted blood using soRT-FDC. The video presents a screen capture of the sorting software window taken during the experiment. The gate of brightness (75–78) and cross-sectional area (56–100 µm2) were set to select for the neutrophils (Fig. 2g,h). The top panel presents an image of the sorting chip. The scale marks in the lower right are spaced 20 µm apart. The analysis ROI is marked with a white rectangle. The SSAW sorting region as well as the target and default outlets are indicated with text labels. Before the sorting start at around 4 s, all cells are directed into the default outlet. Upon sorting start, some of the cells are deflected towards the target outlet. Below the chip image, the bright-field images of cells taken in the ROI are shown only for cells classified for sorting. Next to it, the thresholded and binarized images of all detected cells are displayed. The control panel on the lower left shows several real-time metrics, including total number of sorted cells (Cells sorted), total number of detected cells (Cells detected), the number of detected cells per second (Cells detected [1/s]) and the number of sorted cells per second (Cells sorted [1/s]). The real-time scatter plot of brightness versus deformation of all detected cells is shown on the right-hand side and restarted after ca. 500 events. This screen capture was recorded with VLC media player at approximately 25 frames per s, 100 times lower than the frame rate used for analyzing the cells. This experiment was performed independently three times with similar results, and the screen capture was performed once.
Source data
Source Data Fig. 1
Source data for scatter plots/histograms
Source Data Fig. 2
Source data for scatter plots/histograms
Source Data Fig. 3
Source data for scatter plots/histograms
Source Data Extended Data Fig. 2
Source data for scatter plots/histograms
Source Data Extended Data Fig. 3
Source data for scatter plots/histograms
Source Data Extended Data Fig. 4
Source data for scatter plots/histograms
Source Data Extended Data Fig. 5
Source data for scatter plots/histograms
Source Data Extended Data Fig. 6
Source data for scatter plots
Source Data Extended Data Fig. 7
Source data for box plots
Source Data Extended Data Fig. 8
Source data for scatter plots
Rights and permissions
About this article
Cite this article
Nawaz, A.A., Urbanska, M., Herbig, M. et al. Intelligent image-based deformation-assisted cell sorting with molecular specificity. Nat Methods 17, 595–599 (2020). https://doi.org/10.1038/s41592-020-0831-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-020-0831-y
This article is cited by
-
Classification of fetal and adult red blood cells based on hydrodynamic deformation and deep video recognition
Biomedical Microdevices (2024)
-
Computer vision meets microfluidics: a label-free method for high-throughput cell analysis
Microsystems & Nanoengineering (2023)
-
Microsecond cell triple-sorting enabled by multiple pulse irradiation of femtosecond laser
Scientific Reports (2023)
-
Rapid deformability cytometry for tissue biopsies
Nature Biomedical Engineering (2023)
-
A method for real-time mechanical characterisation of microcapsules
Biomechanics and Modeling in Mechanobiology (2023)