Abstract
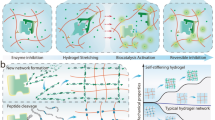
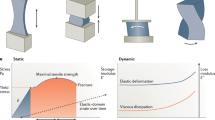
We developed entangled link-augmented stretchable tissue-hydrogel (ELAST), a technology that transforms tissues into elastic hydrogels to enhance macromolecular accessibility and mechanical stability simultaneously. ELASTicized tissues are highly stretchable and compressible, which enables reversible shape transformation and faster delivery of probes into intact tissue specimens via mechanical thinning. This universal platform may facilitate rapid and scalable molecular phenotyping of large-scale biological systems, such as human organs.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
All data supporting the findings of this study are included in figures and videos as representative images or data points in the plots. Additional images other than the representative images are available from the corresponding author upon reasonable request. Resources that may help enable general users to establish the methodology are freely available online (http://www.chunglabresources.org).
Code availability
The custom code used in this study is available from the corresponding author upon reasonable request.
References
Chung, K. et al. Structural and molecular interrogation of intact biological systems. Nature 497, 332–337 (2013).
Chen, F., Tillberg, P. W. & Boyden, E. S. Expansion microscopy. Science 347, 543–548 (2015).
Sylwestrak, E. L., Rajasethupathy, P., Wright, M. A., Jaffe, A. & Deisseroth, K. Multiplexed intact-tissue transcriptional analysis at cellular resolution. Cell 164, 792–804 (2016).
Ku, T. et al. Multiplexed and scalable super-resolution imaging of three-dimensional protein localization in size-adjustable tissues. Nat. Biotechnol. 34, 973–981 (2016).
Wang, X. et al. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science 361, eaat5691 (2018).
Ueda, H. R. et al. Tissue clearing and its applications in neuroscience. Nat. Rev. Neurosci. 21, 61–79 (2020).
Chen, F. et al. Nanoscale imaging of RNA with expansion microscopy. Nat. Methods 13, 679–684 (2016).
Park, H.-E. et al. Scalable and isotropic expansion of tissues with simply tunable expansion ratio. Adv. Sci. 6, 1901673 (2019).
Zhang, Y. S. & Khademhosseini, A. Advances in engineering hydrogels. Science 356, eaaf3627 (2017).
Okumura, Y. & Ito, K. The polyrotaxane gel: a topological gel by figure-of-eight cross-links. Adv. Mater. 13, 485–487 (2001).
Zhong, M. J., Wang, R., Kawamoto, K., Olsen, B. D. & Johnson, J. A. Quantifying the impact of molecular defects on polymer network elasticity. Science 353, 1264–1268 (2016).
Gong, J. P., Katsuyama, Y., Kurokawa, T. & Osada, Y. Double-network hydrogels with extremely high mechanical strength. Adv. Mater. 15, 1155–1158 (2003).
Sun, J.-Y. et al. Highly stretchable and tough hydrogels. Nature 489, 133–136 (2012).
Gong, J. P. Materials both tough and soft. Science 344, 161–162 (2014).
Ball, R. C., Doi, M., Edwards, S. F. & Warner, M. Elasticity of entangled networks. Polymer 22, 1010–1018 (1981).
Hama, H. et al. Scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat. Neurosci. 14, 1481–1488 (2011).
Ke, M.-T., Fujimoto, S. & Imai, T. SeeDB: a simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction. Nat. Neurosci. 16, 1154–1161 (2013).
Pan, C. et al. Shrinkage-mediated imaging of entire organs and organisms using uDISCO. Nat. Methods 13, 859–867 (2016).
Murakami, T. C. et al. A three-dimensional single-cell-resolution whole-brain atlas using CUBIC-X expansion microscopy and tissue clearing. Nat. Neurosci. 21, 625–637 (2018).
Murray, E. et al. Simple, scalable proteomic imaging for high-dimensional profiling of intact systems. Cell 163, 1500–1514 (2015).
Zhao, S. et al. Cellular and molecular probing of intact human organs. Cell 180, 796–812 (2020).
Kubota, S. I. et al. Whole-body profiling of cancer metastasis with single-cell resolution. Cell Rep. 20, 236–250 (2017).
Park, Y.-G. et al. Protection of tissue physicochemical properties using polyfunctional crosslinkers. Nat. Biotechnol. 37, 73–83 (2019).
Lancaster, M. A. & Knoblich, J. A. Generation of cerebral organoids from human pluripotent stem cells. Nat. Protoc. 9, 2329–2340 (2014).
Mellios, N. et al. MeCP2-regulated miRNAs control early human neurogenesis through differential effects on ERK and AKT signaling. Mol. Psychiatr. 23, 1051–1065 (2018).
Acknowledgements
We thank the entire Chung Laboratory for support and discussions. We thank S.-C. Chen and J. Wang for the development of the vibrating-blade microtome, and D. H. Yun for its operation. We also thank K. Xie for mouse sample preparation, Y. Tian for the initial diffusion modeling work and R. Thorndike-Breeze for comments on improving the manuscript. We thank H. Jung and H. Kim for providing the histology facility and helping the H&E experiment. K.C. was supported by the Burroughs Wellcome Fund Career Awards at the Scientific Interface, the Searle Scholars Program, the Packard Award in Science and Engineering, the NARSAD Young Investigator Award, the Institute for Basic Science grant no. IBS-R026-D1 and the McKnight Foundation Technology Award. W.G. was supported by the National Science Foundation Graduate Research Fellowship under grant no. 1122374. A.A. was supported by the JPB Foundation (Picower Fellowship). M.P.F. was partially supported by NIA grant no. P50 AG005134. This work was supported by the JPB Foundation (grant nos. PIIF and PNDRF), the NCSOFT Cultural Foundation, and the NIH (grant nos. 1-DP2-ES027992, U01MH117072).
Author information
Authors and Affiliations
Contributions
T.K. and K.C. designed the experiments and wrote the paper with input from other authors. T.K. developed ELAST and conducted hydrogel experiments, evaluation of mechanical properties and tissue deformation, antibody screening and delivery tests, and the deep-tissue labeling experiment. W.G. modeled the diffusion profiles in hydrogels and conducted the cyclic compression experiment to assess depth-wise immunostaining quality. N.B.E., T.K. and W.G. developed and operated the cyclic compression device. N.B.E. formulated the RI-matching media. C.H.S. and T.K. established the mouse SHIELD protocol for ELAST. C.H.S. conducted the in situ hybridization experiment. A.A. and W.G. conducted the organoid experiment. J.-G.K. and T.K. conducted the downstream histology experiment. M.P.F. provided the human brain tissue specimens. K.C. supervised all aspects of the work.
Corresponding author
Ethics declarations
Competing interests
The ELAST concepts and applications are covered in a pending patent application owned by MIT (K.C. and T.K.). K.C. is a cofounder of LifeCanvas Technologies, a startup that provides solutions for 3D tissue processing.
Additional information
Peer review information Nina Vogt was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Dense entangled pAAm hydrogels that are elastic, tough, and stretchable.
a, High-concentration acrylamide (AAm) solution forms an entangled hydrogel in the absence of cross-linker, N, \(N^\prime\)-methylenebisacrylamide (MBAA). b, A compression test on 40% (wt/vol) pAAm hydrogels with (upper panels) and without (lower panels) 0.5% (wt/vol) MBAA. The gels were prepared with 0.005% (wt/vol) VA-044 and nitrogen gas-bubbling. Gels with 9-mm thickness were compressed to 1 mm. λ, compression strain. c, A demonstration of stretching a 36% (wt/vol) entangled pAAm gel (no MBAA and 0.005% (wt/vol) VA-044 with nitrogen gas-bubbling). Representative images are shown from experiments repeated two times (a and b) and three times (c) with similar results.
Extended Data Fig. 2 ELASTicization and compression of various mouse organs and a cerebral organoid.
a, Compression of ELASTicized mouse organs. Samples were manually compressed between two glass plates. The experiment was not repeated. b, Compression of an ELASTicized cerebral organoid. Scale bars, 5 mm. Experiment was repeated two times with similar results.
Extended Data Fig. 3 Downstream histology of ELASTicized tissue.
a, An ELASTicized 1-mm-thick mouse brain slice block was immunolabeled with an anti-NeuN antibody and imaged by confocal fluorescence microscopy. b, The same sample in a was then cryo-sectioned into 10-μm-thick sections. One section was histologically stained with hematoxylin and eosin with following a series of dehydration steps using ethanol solutions and xylene. The image was obtained by a bright-field microscope. Note that the image planes were not intended to match. Scale bar (adjusted to match the original tissue dimensions), 200 μm. The hematoxylin and eosin staining experiment in b was repeated six times with similar results using two NeuN-stained samples as in a.
Supplementary information
Supplementary information
Supplementary Figs. 1–8 and Tables 1–3.
Supplementary Video 1
Stretching of the unswollen 36% (wt/vol) entangled pAAm hydrogel shown in Extended Data Fig. 1c.
Supplementary Video 2
Stretching of the ELASTicized 2-mm-thick coronal human brain hemisphere slab shown in Fig. 2c.
Supplementary Video 3
Stretching of an ELASTicized 1-mm-thick coronal mouse brain hemisphere slice.
Supplementary Video 4
Compression of ELASTicized mouse organs.
Supplementary Video 5
A demonstration of the operation of the cyclic compression device. The cycling was set to 1 mm and 40 s for compression and 10 mm and 10 s for release.
Supplementary Video 6
Axial navigation of the image volumes of the ELASTicized 3-mm-thick mouse brain block in Fig. 3l showing preserved tissue micro-structures after about 1,500 times of repeated sixfold compression for 1 d. A representative image from four repeated experiments included in Fig. 3m.
Supplementary Video 7
Axial navigation of the image volume in Fig. 3n showing full penetration and complete labeling of an ELASTicized 5-mm-thick human brain cortical block with anti-GFAP antibodies. Experiment was not repeated.
Supplementary Video 8
Calretinin-positive interneurons (orange) and NF-L-positive axonal projections (blue) in the human cerebral cortex sample shown in Fig. 3o that was rapidly immunolabeled via sample-thinning. Experiment was repeated two times with similar results.
Supplementary Video 9
Axial navigation of the image volume in Fig. 3o showing complete immunolabeling of calretinin-positive interneurons and NF-L-positive axonal projections. Experiment was repeated two times with similar results.
Rights and permissions
About this article
Cite this article
Ku, T., Guan, W., Evans, N.B. et al. Elasticizing tissues for reversible shape transformation and accelerated molecular labeling. Nat Methods 17, 609–613 (2020). https://doi.org/10.1038/s41592-020-0823-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-020-0823-y
This article is cited by
-
Nasopharyngeal lymphatic plexus is a hub for cerebrospinal fluid drainage
Nature (2024)
-
An end-to-end workflow for nondestructive 3D pathology
Nature Protocols (2024)
-
Whole-body cellular mapping in mouse using standard IgG antibodies
Nature Biotechnology (2024)
-
Three dimensional evaluation of cerebrovascular density and branching in chronic traumatic encephalopathy
Acta Neuropathologica Communications (2023)
-
Expandable ELAST for super-resolution imaging of thick tissue slices using a hydrogel containing charged monomers
Scientific Reports (2023)