Abstract
Behaviors emerge from activity throughout the brain, but noninvasive optical access in adult vertebrate brains is limited. We show that three-photon (3P) imaging through the head of intact adult zebrafish allows structural and functional imaging at cellular resolution throughout the telencephalon and deep into the cerebellum and optic tectum. With 3P imaging, considerable portions of the brain become noninvasively accessible from embryo to sexually mature adult in a vertebrate model.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
The imaging and scattering length data sets that support the findings of this study are available from the corresponding author upon reasonable request.
Code availability
The software codes used for the analyses are available from the corresponding author upon reasonable request.
References
Fetcho, J. R. & O’Malley, D. M. Visualization of active neural circuitry in the spinal-cord of intact zebrafish. J. Neurophysiol. 73, 399–406 (1995).
Cox, K. J. & Fetcho, J. R. Labeling blastomeres with a calcium indicator: a non-invasive method of visualizing neuronal activity in zebrafish. J. Neurosci. Methods 68, 185–191 (1996).
O’Malley, D. M., Kao, Y.-H. & Fetcho, J. R. Imaging the functional organization of zebrafish hindbrain segments during escape behaviors. Neuron 17, 1145–1155 (1996).
Higashijima, S., Masino, M. A., Mandel, G. & Fetcho, J. R. Imaging neuronal activity during zebrafish behavior with a genetically encoded calcium indicator. J. Neurophysiol. 90, 3986–3997 (2003).
Ahrens, M. B. et al. Brain-wide neuronal dynamics during motor adaptation in zebrafish. Nature 485, 471–477 (2012).
Vladimirov, N. et al. Light-sheet functional imaging in fictively behaving zebrafish. Nat. Methods 11, 883–884 (2014).
Andalman, A. S. et al. Neuronal dynamics regulating brain and behavioral state transitions. Cell 177, 970–985 (2019).
Li, J. et al. Early development of functional spatial maps in the zebrafish olfactory bulb. J. Neurosci. 25, 5784–5795 (2005).
Jacobson, G. A., Rupprecht, P. & Friedrich, R. W. Experience-dependent plasticity of odor representations in the telencephalon of zebrafish. Curr. Biol. 28, 1–14 e13 (2018).
Guesmi, K. et al. Dual-color deep-tissue three-photon microscopy with a multiband infrared laser. Light Sci. Appl. 7, 12 (2018).
White, R. M. et al. Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell 2, 183–189 (2008).
Ouzounov, D. G. et al. In vivo three-photon imaging of activity of GCaMP6-labeled neurons deep in intact mouse brain. Nat. Methods 14, 388–390 (2017).
Wang, T. et al. Three-photon imaging of mouse brain structure and function through the intact skull. Nat. Methods 15, 789–792 (2018).
Farrar, M. J., Wise, F. W., Fetcho, J. R. & Schaffer, C. B. In vivo imaging of myelin in the vertebrate central nervous system using third harmonic generation microscopy. Biophys. J. 100, 1362–1371 (2011).
Park, H. C. et al. Analysis of upstream elements in the HuC promoter leads to the establishment of transgenic zebrafish with fluorescent neurons. Dev. Biol. 227, 279–293 (2000).
Satou, C., Kimura, Y. & Higashijima, S. Generation of multiple classes of V0 neurons in zebrafish spinal cord: progenitor heterogeneity and temporal control of neuronal diversity. J. Neurosci. 32, 1771–1783 (2012).
Bae, Y.-K. et al. Anatomy of zebrafish cerebellum and screen for mutations affecting its development. Dev. Biol. 330, 406–426 (2009).
Corbo, C. P., Othman, N. A., Gutkin, M. C., Alonso Adel, C. & Fulop, Z. L. Use of different morphological techniques to analyze the cellular composition of the adult zebrafish optic tectum. Microsc. Res. Tech. 75, 325–333 (2012).
Diotel, N. et al. Comprehensive expression map of transcription regulators in the adult zebrafish telencephalon reveals distinct neurogenic niches. J. Comp. Neurol. 523, 1202–1221 (2015).
Britz, R., Conway, K. W. & Ruber, L. Spectacular morphological novelty in a miniature cyprinid fish, Danionella dracula n. sp. Proc. Biol. Sci. 276, 2179–2186 (2009).
Acknowledgements
The work was supported by grant nos. NSF DBI-1707312, NIH 1U01NS103516, NSF IOS-1457108 and NIH S10OD012287. We thank C. Walcott (Cornell University) for photographic assistance, D. Mclean (Northwestern University) for his drawing skill, M. Farrar (Messiah College) for the mTdTomato construct and T. Porri (Cornell University) for performing the CT scans.
Author information
Authors and Affiliations
Contributions
D.S., K.E.K., D.M.C, D.G.O., N.A., C.X. and J.R.F. contributed to experimental design and conceptual ideas. D.S and D.G.O. built and maintained the three-photon microscopes. K.E.K, D.M.C., D.S., D.G.O. and N.A. performed three-photon imaging experiments of zebrafish. Analysis was conducted by D.M.C., D.S., N.A. and K.E.K. R.T. and N.A. performed Danionella three-photon imaging. A.B. and J.R.F. supervised Danionella imaging and analysis. J.R.F. and D.M.C. prepared the manuscript with assistance from K.E.K. and input from all authors. J.R.F. and C.X. supervised all aspects of the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nina Vogt was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Structural features visible in optical sections with THG that facilitate navigation through the brain.
A. Telencephalic region, showing from left to right: the skin surface features; the transition into the brain as shadows of neurons become visible; and deeper in the telencephalon where the shadows of the neurons clustered at the midline are evident, as are scattered neurons throughout the section. B. Optic Tectum, showing from left to right: axon bundles on the surface of the tectum just below the skin and skull, with scales on the skin visible; deeper axon bundles in fiber layers of the tectum; densely packed shadows of cell bodies deep in the tectum; a major blood vessel lying adjacent to the ventricle deep in tectum. OT N= 16, Tel N=12.
Extended Data Fig. 2 Third harmonic generation images at different depths in the cerebellar and optic tectal areas.
Squares on brain diagrams on the right show approximate areas of imaging, and dashed lines represent approximate brain midlines. a, Cerebellar region from shallow to deep imaging. At 60 μm, some skin is visible (red asterisk) due to the curvature of the brain, as well as several blood vessels (red triangles) under the skin and above the skull. At 120 μm, skull features are evident. Deeper into the brain, fiber tracts predominate (red arrows). b, Optic tectal region from shallow to deep. At 147 μm, some skull is evident (green square) as well as some brain. Tectal fiber tracts are evident (green asterisk) at multiple depths (see 147, 245 and 378 μm) with a thick commissural fiber tract present (green triangle) at 490 μm. At deeper layers fiber tracts predominate (green arrows). CB N=10, OT N=16.
Extended Data Fig. 3 Signal to Background ratio.
Signal-to-background ratio (SBR) in shallow and deep regions in three areas of the brain. a–c, Fluorescence intensity profiles along the lines across GCaMP6s-labeled neuronal nuclei (N=1 each). d–f, Fluorescence intensity profiles along the lines across blood vessels labelled with fluorescein (N=2 each). To produce each plot, a five-pixel-wide line was drawn across the blood vessel or nucleus. The 5 pixel values were averaged to get an intensity value for the corresponding position along the line. The x axis corresponds to the lines shown on the images to the right. Scale bar 50 μm.
Extended Data Fig. 4 3P Axial resolution measurement.
a, 3P image of a small fluorescein-labeled blood vessel (blue arrowhead) imaged about 200 μm deep into the optic tectum. Image stack was taken with 0.3 μm axial steps at 3 µs pixel time (512x512 pixels/frame) and 4 times averaging. b, Intensity profile along the axial direction fitted with a Gaussian curve to characterize the upper bound of axial resolution. The full width at half maximum of the Gaussian fit is 2.9 μm. N=1.
Extended Data Fig. 5 Example photon counts from telencephalic neurons.
Rows a–c, show neurons at different depths. In each panel, the plots in magenta and green show photon counts from two neurons at that depth, with the monitored neurons marked with m and g in the optical section shown at the right. Telencephalon functional imaging N= 9.
Extended Data Fig. 6 Raster plot of photon counts for all of the neurons shown in main Fig. 2.
Neurons were monitored at different depths in: A. telencephalon, B. optic tectum and C. cerebellum. Functional imaging: Tel N = 9, OT N = 6, CB N = 2.
Extended Data Fig. 7 Experimental setup.
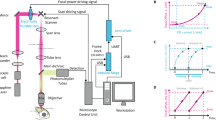
(a) schematic drawing of the excitation sources and the optical path of the imaging setup. OPA – optical parametric oscillator, HWP – Half wave plate, PBS – Polarizing Beam splitter, DM – Dichroic mirror, PMT – Photomultiplier tube. inset(i) - Optical path for the prism compressor for the 1300 nm setup. inset(ii) - Optical path for the silicon plate compressor for the 1700 nm setup.
Supplementary information
Supplementary Information
Supplementary Table 1
Video 1
Blood flow imaged with THG at 8.5 Hz.
Video 2
Video of nonlinear damage induced by excessive laser intensity.
Video 3
Video from a montage of z stacks through the depth of telencephalon on one entire side of the brain from superficial to deep.
Video 4
Three-dimensional video of one half of the telencephalon made from the z stacks shown in Supplementary Video 3.
Rights and permissions
About this article
Cite this article
Chow, D.M., Sinefeld, D., Kolkman, K.E. et al. Deep three-photon imaging of the brain in intact adult zebrafish. Nat Methods 17, 605–608 (2020). https://doi.org/10.1038/s41592-020-0819-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-020-0819-7
This article is cited by
-
Image restoration of degraded time-lapse microscopy data mediated by near-infrared imaging
Nature Methods (2024)
-
Label-free imaging of red blood cells and oxygenation with color third-order sum-frequency generation microscopy
Light: Science & Applications (2023)
-
The emerging vertebrate model species for neurophysiological studies is Danionella cerebrum, new species (Teleostei: Cyprinidae)
Scientific Reports (2021)
-
Zebrafish capable of generating future state prediction error show improved active avoidance behavior in virtual reality
Nature Communications (2021)
-
Reinforcing neuron extraction and spike inference in calcium imaging using deep self-supervised denoising
Nature Methods (2021)