Abstract
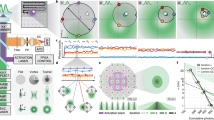
Intracellular diffusion underlies vital cellular processes. However, it remains difficult to elucidate how an unbound protein diffuses inside the cell with good spatial resolution and sensitivity. Here we introduce single-molecule displacement/diffusivity mapping (SMdM), a super-resolution strategy that enables the nanoscale mapping of intracellular diffusivity through local statistics of the instantaneous displacements of freely diffusing single molecules. We thus show that the diffusion of an average-sized protein in the mammalian cytoplasm and nucleus is spatially heterogeneous at the nanoscale, and that variations in local diffusivity correlate with the ultrastructure of the actin cytoskeleton and the organization of the genome, respectively. SMdM of differently charged proteins further unveils that the possession of positive, but not negative, net charges drastically impedes diffusion, and that the rate is determined by the specific subcellular environments. We thus unveil rich heterogeneities and charge effects in intracellular diffusion at the nanoscale.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Code availability
The custom codes for the data analysis used in this study are available from the corresponding author upon request.
References
Kuimova, M. K. et al. Imaging intracellular viscosity of a single cell during photoinduced cell death. Nat. Chem. 1, 69–73 (2009).
Ebbinghaus, S., Dhar, A., McDonald, D. & Gruebele, M. Protein folding stability and dynamics imaged in a living cell. Nat. Methods 7, 319–323 (2010).
Wirth, A. J. & Gruebele, M. Quinary protein structure and the consequences of crowding in living cells: leaving the test-tube behind. Bioessays 35, 984–993 (2013).
Yang, Z. G. et al. Macro-/micro-environment-sensitive chemosensing and biological imaging. Chem. Soc. Rev. 43, 4563–4601 (2014).
Boersma, A. J., Zuhorn, I. S. & Poolman, B. A sensor for quantification of macromolecular crowding in living cells. Nat. Methods 12, 227–229 (2015).
Rivas, G. & Minton, A. P. Macromolecular crowding in vitro, in vivo, and in between. Trends Biochem. Sci. 41, 970–981 (2016).
Lippincott-Schwartz, J., Snapp, E. & Kenworthy, A. Studying protein dynamics in living cells. Nat. Rev. Mol. Cell Biol. 2, 444–456 (2001).
Ishikawa-Ankerhold, H. C., Ankerhold, R. & Drummen, G. P. C. Advanced fluorescence microscopy techniques-FRAP, FLIP, FLAP, FRET and FLIM. Molecules 17, 4047–4132 (2012).
Digman, M. A. & Gratton, E. Lessons in fluctuation correlation spectroscopy. Annu. Rev. Phys. Chem. 62, 645–668 (2011).
Ries, J. & Schwille, P. Fluorescence correlation spectroscopy. Bioessays 34, 361–368 (2012).
Machan, R. & Wohland, T. Recent applications of fluorescence correlation spectroscopy in live systems. FEBS Lett. 588, 3571–3584 (2014).
Krieger, J. W. et al. Imaging fluorescence (cross-) correlation spectroscopy in live cells and organisms. Nat. Protoc. 10, 1948–1974 (2015).
Enderlein, J., Gregor, I., Patra, D., Dertinger, T. & Kaupp, U. B. Performance of fluorescence correlation spectroscopy for measuring diffusion and concentration. ChemPhysChem 6, 2324–2336 (2005).
Eggeling, C. et al. Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature 457, 1159–1162 (2009).
Sezgin, E. et al. Measuring nanoscale diffusion dynamics in cellular membranes with super-resolution STED-FCS. Nat. Protoc. 14, 1054–1083 (2019).
Manley, S. et al. High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nat. Methods 5, 155–157 (2008).
Chenouard, N. et al. Objective comparison of particle tracking methods. Nat. Methods 11, 281–289 (2014).
Kusumi, A., Tsunoyama, T. A., Hirosawa, K. M., Kasai, R. S. & Fujiwara, T. K. Tracking single molecules at work in living cells. Nat. Chem. Biol. 10, 524–532 (2014).
Cognet, L., Leduc, C. & Lounis, B. Advances in live-cell single-particle tracking and dynamic super-resolution imaging. Curr. Opin. Chem. Biol. 20, 78–85 (2014).
Manzo, C. & Garcia-Parajo, M. F. A review of progress in single particle tracking: from methods to biophysical insights. Rep. Prog. Phys. 78, 124601 (2015).
Elf, J. & Barkefors, I. Single-molecule kinetics in living cells. Annu. Rev. Biochem. 88, 635–659 (2019).
Milo, R. & Phillips, R. Cell Biology by the Numbers (Garland Science, 2016).
Elf, J., Li, G. W. & Xie, X. S. Probing transcription factor dynamics at the single-molecule level in a living cell. Science 316, 1191–1194 (2007).
English, B. P. et al. Single-molecule investigations of the stringent response machinery in living bacterial cells. Proc. Natl Acad. Sci. USA 108, E365–E373 (2011).
Rust, M. J., Bates, M. & Zhuang, X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3, 793–795 (2006).
Betzig, E. et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science 313, 1642–1645 (2006).
Hess, S. T., Girirajan, T. P. K. & Mason, M. D. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys. J. 91, 4258–4272 (2006).
Zhang, M. S. et al. Rational design of true monomeric and bright photoactivatable fluorescent proteins. Nat. Methods 9, 727–729 (2012).
Swaminathan, R., Hoang, C. P. & Verkman, A. S. Photobleaching recovery and anisotropy decay of green fluorescent protein GFP-S65T in solution and cells: cytoplasmic viscosity probed by green fluorescent protein translational and rotational diffusion. Biophys. J. 72, 1900–1907 (1997).
Xu, K., Babcock, H. P. & Zhuang, X. Dual-objective STORM reveals three-dimensional filament organization in the actin cytoskeleton. Nat. Methods 9, 185–188 (2012).
Seksek, O., Biwersi, J. & Verkman, A. S. Translational diffusion of macromolecule-sized solutes in cytoplasm and nucleus. J. Cell Biol. 138, 131–142 (1997).
Boisvert, F. M., van Koningsbruggen, S., Navascues, J. & Lamond, A. I. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 8, 574–585 (2007).
Marfori, M. et al. Molecular basis for specificity of nuclear import and prediction of nuclear localization. Biochim. Biophys. Acta Mol. Cell Res. 1813, 1562–1577 (2011).
Xiong, K. & Blainey, P. C. Molecular sled sequences are common in mammalian proteins. Nucleic Acids Res. 44, 2266–2273 (2016).
Potma, E. O. et al. Reduced protein diffusion rate by cytoskeleton in vegetative and polarized Dictyostelium cells. Biophys. J. 81, 2010–2019 (2001).
Baum, M., Erdel, F., Wachsmuth, M. & Rippe, K. Retrieving the intracellular topology from multi-scale protein mobility mapping in living cells. Nat. Commun. 5, 4494 (2014).
Di Rienzo, C., Cardarelli, F., Di Luca, M., Beltram, F. & Gratton, E. Diffusion tensor analysis by two-dimensional pair correlation of fluorescence fluctuations in cells. Biophys. J. 111, 841–851 (2016).
Bancaud, A. et al. Molecular crowding affects diffusion and binding of nuclear proteins in heterochromatin and reveals the fractal organization of chromatin. EMBO J. 28, 3785–3798 (2009).
Dross, N. et al. Mapping eGFP oligomer mobility in living cell nuclei. PLoS ONE 4, e5041 (2009).
Schavemaker, P. E., Smigiel, W. M. & Poolman, B. Ribosome surface properties may impose limits on the nature of the cytoplasmic proteome. eLife 6, e30084 (2017).
Lodish, H. et al. in Molecular Cell Biology Ch. 7, 253 (W.H. Freeman, 2003).
Zustiak, S. P., Nossal, R. & Sackett, D. L. Hindered diffusion in polymeric solutions studied by fluorescence correlation spectroscopy. Biophys. J. 101, 255–264 (2011).
Lawrence, M. S., Phillips, K. J. & Liu, D. R. Supercharging proteins can impart unusual resilience. J. Am. Chem. Soc. 129, 10110–10112 (2007).
Zhang, Z., Kenny, S. J., Hauser, M., Li, W. & Xu, K. Ultrahigh-throughput single-molecule spectroscopy and spectrally resolved super-resolution microscopy. Nat. Methods 12, 935–938 (2015).
Yan, R., Moon, S., Kenny, S. J. & Xu, K. Spectrally resolved and functional super-resolution microscopy via ultrahigh-throughput single-molecule spectroscopy. Acc. Chem. Res. 51, 697–705 (2018).
Anderson, C. M., Georgiou, G. N., Morrison, I. E. G., Stevenson, G. V. W. & Cherry, R. J. Tracking of cell surface receptors by fluorescence digital imaging microscopy using a charge-coupled device camera. Low-density lipoprotein and influenza virus receptor mobility at 4 °C. J. Cell Sci. 101, 415–425 (1992).
Kues, T., Peters, R. & Kubitscheck, U. Visualization and tracking of single protein molecules in the cell nucleus. Biophys. J. 80, 2954–2967 (2001).
Lin, W. C. et al. H-Ras forms dimers on membrane surfaces via a protein-protein interface. Proc. Natl Acad. Sci. USA 111, 2996–3001 (2014).
Hansen, A. S. et al. Robust model-based analysis of single-particle tracking experiments with Spot-On. eLife 7, e33125 (2018).
Acknowledgements
We thank S. Moon for discussion, and M. He and Y. Shyu for help with preparation of the DNA constructs. This work was supported by the National Institute of General Medical Sciences of the National Institutes of Health (grant no. DP2GM132681), the Beckman Young Investigator Program and the Packard Fellowships for Science and Engineering to K.X. K.X. is a Chan Zuckerberg Biohub investigator.
Author information
Authors and Affiliations
Contributions
K.X. conceived the research. L.X. and K.C. designed and conducted the experiments. All authors contributed to experimental designs, data analysis and paper writing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 SMdM results at different single-molecule densities.
Free mEos3.2 was expressed in the cytoplasm of a PtK2 cell, and SMdM was performed on the same cell at a low single-molecule density of ~0.05 molecules/µm2/frame for 60,000 pairs of pulses (a–c), or at a high single-molecule density of ~0.11 molecules/µm2/frame for 30,000 pairs of pulses (d–f) by increasing the power of the photoactivation (405 nm) laser. (a) SMdM diffusivity map for the low single-molecule density experiment, obtained by spatially binning the single-molecule displacement d data onto 120×120 nm2 grids, and then individually fitting the distribution of d in each bin to Eq. 2 through MLE. (b, c) Distribution of 1-ms single-molecule displacement for two 360×360 nm2 areas inside (b; white arrow in a) and outside (c; red arrow in a) a linear structure of reduced local diffusivity, respectively. Blue lines are MLE results using Eq. 2, with resultant D and uncertainty σ labeled in each panel. (d–f) Results of the high single-molecule density experiment: comparable D values are obtained with the much-reduced number of pulse pairs, despite an increased background due to single-molecule mismatch. These experiments were independently repeated 10 times with similar results.
Extended Data Fig. 2 Additional SMdM results of free mEos3.2 in the cytoplasm of live U2OS and PtK2 cells, and correlated SMLM of the actin cytoskeleton.
(a, b) Correlated SMdM diffusivity map for a live U2OS cell (a) vs. SMLM image of Alexa Fluor 647 phalloidin-labeled actin in the fixed cell (b). (a) and (b) were independently repeated 4 times with similar results. (c, d) Additional SMdM diffusivity maps for the cytoplasm of PtK2 cells. (c) and (d) were independently repeated 11 times with similar results.
Extended Data Fig. 3 Additional SMdM results of free mEos3.2 in the nuclei of live PtK2 cells, and correlated SMLM of DNA.
(a, d, f) SMdM diffusivity maps of 3 different cells. (b) Bright-field transmission image of the same view as (a), visualizing the nucleolus. (c, e, g) SMLM images of the fixed cells in (a, d, f) using the DNA stain NucSpot Live 650. We note that as the SMLM of DNA was performed after fixation and multiple washing steps, it was difficult to image at exactly the same focal plane as the live-cell SMdM experiment, which accounts for some of the apparent structural mismatches. Scale bars in all panels: 2 µm. These experiments were independently repeated 23 times with similar results.
Extended Data Fig. 4 SMdM of mEos3.2-NLS and correlated SMLM of DNA.
(a) SMdM diffusivity map of mEos3.2-NLS in the nucleus of a live PtK2 cell. (b) SMLM image of the fixed cell using the DNA stain NucSpot Live 650. (c) Overlay of (a) and (b). Scale bars: 2 µm. These experiments were independently repeated 18 times with similar results.
Extended Data Fig. 5 SMdM of free mEos3.2 using 488 nm excitation without photoactivation.
(a, b) SMdM diffusivity maps of mEos3.2-C1 in the cytoplasm of live PtK2 cells, obtained by exciting the un-photoconverted, “green” form of mEos3.2 single molecules with 488 nm excitation. (a) and (b) were independently repeated 6 times with similar results. (c, d) Distribution of 1-ms single-molecule displacement for two 360×360 nm2 areas inside (c; white arrow in a) and outside (d; red arrow in a) a linear structure of reduced local diffusivity, respectively. Blue lines are MLE results using Eq. 2, with resultant D and uncertainty σ labeled in each panel.
Extended Data Fig. 6 Asymmetric effects of negative and positive net charges on intracellular diffusion.
(a) A negatively charged diffuser is readily neutralized by the abundant, small metal cations inside the cell, and so diffuses similarly as neutral counterparts. (b) A positively charged diffuser is not effectively neutralized/screened by the very limited amount of intracellular small anions; its dynamic interactions with the negatively charged, large biomolecules insides the cell substantially hinder diffusion.
Supplementary Information
Supplementary Information
Supplementary Tables 1 and 2.
Rights and permissions
About this article
Cite this article
Xiang, L., Chen, K., Yan, R. et al. Single-molecule displacement mapping unveils nanoscale heterogeneities in intracellular diffusivity. Nat Methods 17, 524–530 (2020). https://doi.org/10.1038/s41592-020-0793-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-020-0793-0
This article is cited by
-
Geometric deep learning reveals the spatiotemporal features of microscopic motion
Nature Machine Intelligence (2023)
-
Super-resolving microscopy reveals the localizations and movement dynamics of stressosome proteins in Listeria monocytogenes
Communications Biology (2023)
-
Machine-learning-powered extraction of molecular diffusivity from single-molecule images for super-resolution mapping
Communications Biology (2023)
-
Spatially mapping the diffusivity of proteins in live cells based on cumulative area analysis
Science China Chemistry (2023)
-
Stress induced TDP-43 mobility loss independent of stress granules
Nature Communications (2022)