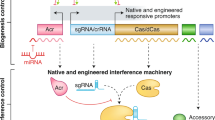
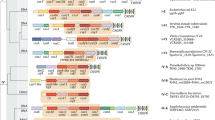
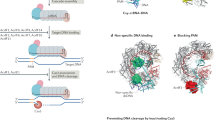
Abstract
Clustered, regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated (Cas) genes, a diverse family of prokaryotic adaptive immune systems, have emerged as a biotechnological tool and therapeutic. The discovery of protein inhibitors of CRISPR-Cas systems, called anti-CRISPR (Acr) proteins, enables the development of more controllable and precise CRISPR-Cas tools. Here we discuss applications of Acr proteins for post-translational control of CRISPR-Cas systems in prokaryotic and mammalian cells, organisms and ecosystems.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Adli, M. The CRISPR tool kit for genome editing and beyond. Nat. Commun. 9, 1911 (2018).
Hsu, P. D., Lander, E. S. & Zhang, F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 157, 1262–1278 (2014).
Hsu, P. D. et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31, 827–832 (2013).
Lee, H. & Kim, J.-S. Unexpected CRISPR on-target effects. Nat. Biotechnol. 36, 703–704 (2018).
Ihry, R. J. et al. p53 inhibits CRISPR-Cas9 engineering in human pluripotent stem cells. Nat. Med. 24, 939–946 (2018).
Li, C. et al. HDAd5/35++ Adenovirus vector expressing anti-CRISPR peptides decreases CRISPR/Cas9 toxicity in human hematopoietic stem cells. Mol. Ther. Methods Clin. Dev. 9, 390–401 (2018). This study demonstrated that acr genes delivered into cells ex vivo can reduce Cas9-associated cytotoxicity and improve engraftment outcomes.
Chew, W. L. et al. A multifunctional AAV-CRISPR-Cas9 and its host response. Nat. Methods 13, 868–874 (2016).
Wang, D. et al. Adenovirus-mediated somatic genome editing of Pten by CRISPR/Cas9 in mouse liver in spite of Cas9-specific immune responses. Hum. Gene Ther. 26, 432–442 (2015).
Borges, A. L., Davidson, A. R. & Bondy-Denomy, J. The discovery, mechanisms, and evolutionary impact of anti-CRISPRs. Annu. Rev. Virol. 4, 37–59 (2017).
Stanley, S. Y. & Maxwell, K. L. Phage-encoded anti-CRISPR defenses. Annu. Rev. Genet. 52, 445–464 (2018).
Trasanidou, D. et al. Keeping CRISPR in check: diverse mechanisms of phage-encoded anti-CRISPRs. FEMS Microbiol. Lett. 366, 1709 (2019).
Knott, G. J. et al. Broad-spectrum enzymatic inhibition of CRISPR-Cas12a. Nat. Struct. Mol. Biol. 26, 315–321 (2019).
Dong, L. et al. An anti-CRISPR protein disables type V Cas12a by acetylation. Nat. Struct. Mol. Biol. 26, 308–314 (2019).
Pawluk, A., Davidson, A. R. & Maxwell, K. L. Anti-CRISPR: discovery, mechanism and function. Nat. Rev. Microbiol. 16, 12–17 (2018).
Bondy-Denomy, J. et al. Multiple mechanisms for CRISPR-Cas inhibition by anti-CRISPR proteins. Nature 526, 136–139 (2015). This study identified multiple mechanisms of inhibition via direct interactions with Cas proteins for the first discovered Acr proteins.
Dong, D. et al. Structural basis of CRISPR-SpyCas9 inhibition by an anti-CRISPR protein. Nature 546, 436–439 (2017). This work identified the mechanism and structure of a Cas9 inhibitor, showing AcrIIA4 binds the PAM-interacting motif of Cas9.
Jiang, F. et al. Temperature-responsive competitive inhibition of CRISPR-Cas9. Mol. Cell 73, 601–610.e5 (2019).
Harrington, L. B. et al. A broad-spectrum inhibitor of CRISPR-Cas9. Cell 170, 1224–1233.e15 (2017).
Bondy-Denomy, J. et al. A unified resource for tracking anti-CRISPR names. CRISPR J. 1, 304–305 (2018).
Rauch, B. J. et al. Inhibition of CRISPR-Cas9 with bacteriophage proteins. Cell 168, 150–158.e10 (2017). This study reported Acr proteins that inhibit SpyCas9 and demonstrated the efficacy of AcrIIA2 and AcrIIA4 in human cells.
Pawluk, A. et al. Naturally occurring off-switches for CRISPR-Cas9. Cell 167, 1829–1838.e9 (2016). This study identified the Acr proteins that inhibit NmeCas9 and demonstrated their efficacy in human cells.
Pickar-Oliver, A. & Gersbach, C. A. The next generation of CRISPR-Cas technologies and applications. Nat. Rev. Mol. Cell Biol. 20, 490–507 (2019).
Choi, K. R. & Lee, S. Y. CRISPR technologies for bacterial systems: Current achievements and future directions. Biotechnol. Adv. 34, 1180–1209 (2016).
Jiang, Y. et al. Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl. Environ. Microbiol. 81, 2506–2514 (2015).
Luo, M. L., Leenay, R. T. & Beisel, C. L. Current and future prospects for CRISPR-based tools in bacteria. Biotechnol. Bioeng. 113, 930–943 (2016).
Makarova, K. S. et al. An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 13, 722–736 (2015).
Jiang, W., Bikard, D., Cox, D., Zhang, F. & Marraffini, L. A. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 31, 233–239 (2013).
Luo, M. L., Mullis, A. S., Leenay, R. T. & Beisel, C. L. Repurposing endogenous type I CRISPR-Cas systems for programmable gene repression. Nucleic Acids Res. 43, 674–681 (2015).
van Belkum, A. et al. Phylogenetic distribution of CRISPR-Cas systems in antibiotic-resistant Pseudomonas aeruginosa. MBio 6, e01796–15 (2015).
Mayo-Muñoz, D. et al. Anti-CRISPR-based and CRISPR-based genome editing of Sulfolobus islandicus Rod-Shaped Virus 2. Viruses 10, 695 (2018). This study demonstrated the use of Acr proteins as selectable markers in viral genome engineering.
Qi, L. S. et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183 (2013).
Pawluk, A. et al. Disabling a type I-E CRISPR-Cas nuclease with a bacteriophage-encoded anti-CRISPR protein. MBio 8, 43 (2017).
Louwen, R., Staals, R. H. J., Endtz, H. P., van Baarlen, P. & van der Oost, J. The role of CRISPR-Cas systems in virulence of pathogenic bacteria. Microbiol. Mol. Biol. Rev. 78, 74–88 (2014).
Nobrega, F. L., Costa, A. R., Kluskens, L. D. & Azeredo, J. Revisiting phage therapy: new applications for old resources. Trends Microbiol. 23, 185–191 (2015).
Muñoz, I. V., Sarrocco, S., Malfatti, L., Baroncelli, R. & Vannacci, G. CRISPR-Cas for fungal genome editing: a new tool for the management of plant diseases. Front. Plant Sci. 10, 135 (2019).
Langner, T., Kamoun, S. & Belhaj, K. CRISPR Crops: plant genome editing toward disease resistance. Annu. Rev. Phytopathol. 56, 479–512 (2018).
Jinek, M. et al. RNA-programmed genome editing in human cells. Elife 2, e00471 (2013).
Mali, P. et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013).
Swarts, D. C. & Jinek, M. Cas9 versus Cas12a/Cpf1: Structure-function comparisons and implications for genome editing. Wiley Interdiscip. Rev. RNA 9, e1481 (2018).
Yao, R. et al. CRISPR-Cas9/Cas12a biotechnology and application in bacteria. Synth. Syst. Biotechnol. 3, 135–149 (2018).
Kleinstiver, B. P. et al. Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells. Nat. Biotechnol. 34, 869–874 (2016).
Kim, D. et al. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat. Biotechnol. 34, 863–868 (2016).
Kim, S., Kim, D., Cho, S. W., Kim, J. & Kim, J. S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 24, 1012–1019 (2014).
Lin, S., Staahl, B. T., Alla, R. K. & Doudna, J. A. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. Elife 3, e04766 (2014).
Nihongaki, Y., Kawano, F., Nakajima, T. & Sato, M. Photoactivatable CRISPR-Cas9 for optogenetic genome editing. Nat. Biotechnol. 33, 755–760 (2015).
Senturk, S. et al. Rapid and tunable method to temporally control gene editing based on conditional Cas9 stabilization. Nat. Commun. 8, 14370 (2017).
Kleinstiver, B. P. et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490–495 (2016).
Slaymaker, I. M. et al. Rationally engineered Cas9 nucleases with improved specificity. Science 351, 84–88 (2016).
Chen, J. S. et al. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature 550, 407–410 (2017).
Shin, J. et al. Disabling Cas9 by an anti-CRISPR DNA mimic. Sci. Adv. 3, e1701620 (2017). This study demonstrated that AcrIIA4 can reduce off-target editing while maintaining on-target editing in human cells.
Yang, S., Li, S. & Li, X.-J. Shortening the half-life of Cas9 maintains its gene editing ability and reduces neuronal toxicity. Cell Rep. 25, 2653–2659.e3 (2018).
Maeder, M. L. et al. Development of a gene-editing approach to restore vision loss in Leber congenital amaurosis type 10. Nat. Med. 25, 229–233 (2019).
Lee, J. et al. Tissue-restricted genome editing in vivo specified by microRNA-repressible anti-CRISPR proteins. RNA rna.071704.119 (2019). This study demonstrated Cas9 inhibition with Acr proteins in mice.
Liu, X. S. et al. Rescue of fragile X syndrome neurons by DNA methylation editing of the FMR1 gene. Cell 172, 979–992.e6 (2018).
Chen, B. et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 155, 1479–1491 (2013).
Wu, X., Mao, S., Ying, Y., Krueger, C. J. & Chen, A. K. Progress and challenges for live-cell imaging of genomic loci using CRISPR-based platforms. Genomics Proteomics Bioinformatics 17, 119–128 (2019).
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A. & Liu, D. R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016).
Gaudelli, N. M. et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 551, 464–471 (2017).
Li, X. et al. Base editing with a Cpf1-cytidine deaminase fusion. Nat. Biotechnol. 36, 324–327 (2018).
Zuo, E. et al. Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science 364, 289–292 (2019).
Jin, S. et al. Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science 364, 292–295 (2019).
Li, J., Xu, Z., Chupalov, A. & Marchisio, M. A. Anti-CRISPR-based biosensors in the yeast S. cerevisiae. 1–14 (2018).
Nakamura, M. et al. Anti-CRISPR-mediated control of gene editing and synthetic circuits in eukaryotic cells. Nat. Commun. 10, 194 (2019). This study demonstrates many applications of Acr proteins in eukaryotic cells, including ‘write protecting’ cells from further editing, CRISPR-based gene regulation circuits, and ligand-inducible AcrIIA4.
Dow, L. E. et al. Inducible in vivo genome editing with CRISPR-Cas9. Nat. Biotechnol. 33, 390–394 (2015).
Hemphill, J., Borchardt, E. K., Brown, K., Asokan, A. & Deiters, A. Optical control of CRISPR/Cas9 gene editing. J. Am. Chem. Soc. 137, 5642–5645 (2015).
Maji, B. et al. A high-throughput platform to identify small-molecule inhibitors of CRISPR-Cas9. Cell 177, 1067–1079.e19 (2019).
Marino, N. D. et al. Discovery of widespread type I and type V CRISPR-Cas inhibitors. Science 362, 240–242 (2018).
Hoffmann, M. D. et al. Cell-specific CRISPR-Cas9 activation by microRNA-dependent expression of anti-CRISPR proteins. Nucleic Acids Res. 47, e75 (2019).
Bubeck, F. et al. Engineered anti-CRISPR proteins for optogenetic control of CRISPR-Cas9. Nat. Methods 15, 924–927 (2018). This study reported an optogenetic AcrIIA4 variant that can be inactivated in cells using light.
Stanley, S. Y. et al. Anti-CRISPR-associated proteins are crucial repressors of anti-CRISPR transcription. Cell 178, 1452–1464.e13 (2019).
Hirosawa, M., Fujita, Y. & Saito, H. Cell-type-specific CRISPR activation with microRNA-responsive AcrllA4 switch. ACS Synth. Biol. 8, 1575–1582 (2019).
Burt, A. Site-specific selfish genes as tools for the control and genetic engineering of natural populations. Proc. Biol. Sci. 270, 921–928 (2003).
Gantz, V. M. et al. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc. Natl Acad. Sci. USA 112, E6736–E6743 (2015).
Hammond, A. et al. A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nat. Biotechnol. 34, 78–83 (2016).
Esvelt, K. M., Smidler, A. L., Catteruccia, F. & Church, G. M. Concerning RNA-guided gene drives for the alteration of wild populations. Elife 3, 20131071 (2014).
Akbari, O. S. et al. BIOSAFETY. Safeguarding gene drive experiments in the laboratory. Science 349, 927–929 (2015).
Basgall, E. M. et al. Gene drive inhibition by the anti-CRISPR proteins AcrIIA2 and AcrIIA4 in Saccharomyces cerevisiae. Microbiology 164, 464–474 (2018). This study demonstrated the ability of AcrIIA2 and AcrIIA4 to halt gene drives in yeast.
Johnston, R. K. et al. Use of anti-CRISPR protein AcrIIA4 as a capture ligand for CRISPR/Cas9 detection. Biosens. Bioelectron. 141, 111361 (2019).
Palmer, D. J., Turner, D. L. & Ng, P. Production of CRISPR/Cas9-mediated self-cleaving helper-dependent adenoviruses. Mol. Ther. Methods Clin. Dev. 13, 432–439 (2019).
Smargon, A. A. et al. Cas13b Is a type VI-B CRISPR-associated RNA-guided RNase differentially regulated by accessory proteins Csx27 and Csx28. Mol. Cell 65, 618–630.e7 (2017).
Pawluk, A. et al. Inactivation of CRISPR-Cas systems by anti-CRISPR proteins in diverse bacterial species. Nat. Microbiol. 1, 16085 (2016).
Bondy-Denomy, J., Pawluk, A., Maxwell, K. L. & Davidson, A. R. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature 493, 429–432 (2013). This study identified phage proteins with Acr function.
Guo, T. W. et al. Cryo-EM structures reveal mechanism and inhibition of DNA targeting by a CRISPR-Cas surveillance complex. Cell 171, 414–426.e12 (2017).
He, F. et al. Anti-CRISPR proteins encoded by archaeal lytic viruses inhibit subtype I-D immunity. Nat. Microbiol. 3, 461–469 (2018).
Pawluk, A., Bondy-Denomy, J., Cheung, V. H. W., Maxwell, K. L. & Davidson, A. R. A new group of phage anti-CRISPR genes inhibits the type I-E CRISPR-Cas system of Pseudomonas aeruginosa. MBio 5, e00896 (2014). e00896–e14.
Fuchsbauer, O. et al. Cas9 Allosteric inhibition by the anti-CRISPR protein AcrIIA6. Mol. Cell 76, 922–937.e7 (2019).
Hynes, A. P. et al. Widespread anti-CRISPR proteins in virulent bacteriophages inhibit a range of Cas9 proteins. Nat. Commun. 9, 2919 (2018).
Lee, J. et al. Potent Cas9 inhibition in bacterial and human cells by AcrIIC4 and AcrIIC5 anti-CRISPR proteins. MBio 9, 1239 (2018).
Sun, W. et al. Structures of Neisseria meningitidis Cas9 complexes in catalytically poised and anti-CRISPR-inhibited states. Mol. Cell 76, 938–952.e5 (2019).
Thavalingam, A. et al. Inhibition of CRISPR-Cas9 ribonucleoprotein complex assembly by anti-CRISPR AcrIIC2. Nat. Commun. 10, 2806–2811 (2019).
Zhu, Y. et al. Diverse mechanisms of CRISPR-Cas9 inhibition by type IIC anti-CRISPR proteins. Mol. Cell 74, 296–309.e7 (2019).
Forsberg, K. J. et al. Functional metagenomics-guided discovery of potent Cas9 inhibitors in the human microbiome. Elife 8, 1709 (2019).
Ka, D., An, S. Y., Suh, J.-Y. & Bae, E. Crystal structure of an anti-CRISPR protein, AcrIIA1. Nucleic Acids Res. 46, 485–492 (2018).
Hynes, A. P. et al. An anti-CRISPR from a virulent streptococcal phage inhibits Streptococcus pyogenes Cas9. Nat. Microbiol. 2, 1374–1380 (2017).
Uribe, R. V. et al. Discovery and characterization of Cas9 inhibitors disseminated across seven bacterial phyla. Cell Host Microbe 25, 233–241.e5 (2019).
Bhoobalan-Chitty, Y., Johansen, T. B., Di Cianni, N. & Peng, X. Inhibition of type III CRISPR-Cas immunity by an archaeal virus-encoded anti-CRISPR protein. Cell 179, 448–458.e11 (2019).
Zhang, H. et al. Structural basis for the inhibition of CRISPR-Cas12a by anti-CRISPR proteins. Cell Host Microbe 25, 815–826.e4 (2019).
Watters, K. E., Fellmann, C., Bai, H. B., Ren, S. M. & Doudna, J. A. Systematic discovery of natural CRISPR-Cas12a inhibitors. Science 9, eaau5138 (2018).
Wandera, K. G. et al. An enhanced assay to characterize anti-CRISPR proteins using a cell-free transcription-translation system. Methods https://doi.org/10.1016/j.ymeth.2019.05.014 (2019).
Acknowledgements
We thank M. Pinilla-Redondo, who made the figures for this manuscript. Acr research in the Bondy-Denomy lab was supported by the University of California San Francisco Program for Breakthrough in Biomedical Research, funded in part by the Sandler Foundation, by an NIH Office of the Director Early Independence Award DP5-OD021344, by NIH R01GM127489, and by DARPA HR0011-17-2-0043. N.D.M. was supported by NIH F32GM133127, B.C. was supported by the Eötvös National Scholarship of Hungary and a Marie Skłodowska-Curie Actions Individual Global Fellowship (number 844093) of the Horizon 2020 Research Program of the European Commission. R.P.R. was funded by Joint Programming Initiative-Antimicrobial Resistance (JIP-AMR; DARWIN project, #7044-00004B), the Innovation Fund Denmark (Trojan Horse Project, #5157-00005B).
Author information
Authors and Affiliations
Contributions
N.D.M. wrote the sections on the applications of Acr proteins for eukaryotic and in vitro systems and regulation of Acr proteins, and the informational boxes on the advantages and limitations of Acr proteins. R.P.R. wrote the introduction, informational box for Acr protein discovery, and Table 1. B.C. wrote the sections on controlling gene drives with Acr proteins and applications of Acr proteins for prokaryotic systems. J.B.-D. supervised and wrote the manuscript with N.D.M, R.P.R. and B.C. All authors contributed to figure content and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
J.B.-D. is a scientific advisory board member of SNIPR Biome and Excision Biotherapeutics and a scientific advisory board member and co-founder of Acrigen Biosciences. J.B.-D. and N.D.M. have filed patents on technology related to anti-CRISPR proteins. R.P.R. is a consultant for Ancilia Inc.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Marino, N.D., Pinilla-Redondo, R., Csörgő, B. et al. Anti-CRISPR protein applications: natural brakes for CRISPR-Cas technologies. Nat Methods 17, 471–479 (2020). https://doi.org/10.1038/s41592-020-0771-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-020-0771-6
This article is cited by
-
Discovery of a high-performance phage-derived promoter/repressor system for probiotic lactobacillus engineering
Microbial Cell Factories (2024)
-
The zoonotic pathogen Wohlfahrtiimonas chitiniclastica – current findings from a clinical and genomic perspective
BMC Microbiology (2024)
-
Anti-CRISPR Anopheles mosquitoes inhibit gene drive spread under challenging behavioural conditions in large cages
Nature Communications (2024)
-
Inhibitors of bacterial immune systems: discovery, mechanisms and applications
Nature Reviews Genetics (2024)
-
Structure-guided discovery of anti-CRISPR and anti-phage defense proteins
Nature Communications (2024)