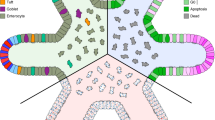
Abstract
Despite the widespread adoption of organoids as biomimetic tissue models, methods to comprehensively analyze cell-type-specific post-translational modification (PTM) signaling networks in organoids are absent. Here, we report multivariate single-cell analysis of such networks in organoids and organoid cocultures. Simultaneous analysis by mass cytometry of 28 PTMs in >1 million single cells derived from small intestinal organoids reveals cell-type- and cell-state-specific signaling networks in stem, Paneth, enteroendocrine, tuft and goblet cells, as well as enterocytes. Integrating single-cell PTM analysis with thiol-reactive organoid barcoding in situ (TOBis) enables high-throughput comparison of signaling networks between organoid cultures. Cell-type-specific PTM analysis of colorectal cancer organoid cocultures reveals that shApc, KrasG12D and Trp53R172H cell-autonomously mimic signaling states normally induced by stromal fibroblasts and macrophages. These results demonstrate how standard mass cytometry workflows can be modified to perform high-throughput multivariate cell-type-specific signaling analysis of healthy and cancerous organoids.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Clevers, H. Modeling development and disease with organoids. Cell 165, 1586–1597 (2016).
Pastula, A. et al. Three-dimensional gastrointestinal organoid culture in combination with nerves or fibroblasts: a method to characterize the gastrointestinal stem cell niche. Stem Cells Int. 2016, 3710836 (2016).
Dijkstra, K. K. et al. Generation of tumor-reactive T cells by co-culture of peripheral blood lymphocytes and tumor organoids. Cell 174, 1586–1598.e12 (2018).
Tuveson, D. & Clevers, H. Cancer modeling meets human organoid technology. Science 364, 952–955 (2019).
Pawson, T. & Scott, J. D. Protein phosphorylation in signaling—50 years and counting. Trends Biochem. Sci. 30, 286–290 (2005).
Miller-Jensen, K., Janes, K. A., Brugge, J. S. & Lauffenburger, D. A. Common effector processing mediates cell-specific responses to stimuli. Nature 448, 604–608 (2007).
Tape, C. J. et al. Oncogenic KRAS regulates tumor cell signaling via stromal reciprocation. Cell 165, 910–920 (2016).
Simmons, A. J. et al. Cytometry-based single-cell analysis of intact epithelial signaling reveals MAPK activation divergent from TNF-ɑ-induced apoptosis in vivo. Mol. Syst. Biol. 11, 835 (2015).
Tape, C. J. Systems biology analysis of heterocellular signaling. Trends Biotechnol. 34, 627–637 (2016).
Haber, A. L. et al. A single-cell survey of the small intestinal epithelium. Nature 551, 333–339 (2017).
Spitzer, M. H. & Nolan, G. P. Mass cytometry: single cells, many features. Cell 165, 780–791 (2016).
Bendall, S. C. et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 332, 687–696 (2011).
Sato, T. et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 (2009).
Behbehani, G. K., Bendall, S. C., Clutter, M. R., Fantl, W. J. & Nolan, G. P. Single-cell mass cytometry adapted to measurements of the cell cycle. Cytom. A 81, 552–566 (2012).
Fienberg, H. G., Simonds, E. F., Fantl, W. J., Nolan, G. P. & Bodenmiller, B. A platinum-based covalent viability reagent for single-cell mass cytometry. Cytom. A 81, 467–475 (2012).
Yin, X. et al. Niche-independent high-purity cultures of Lgr5+ intestinal stem cells and their progeny. Nat. Methods 11, 106–112 (2014).
Rapsomaniki, M. A. et al. CellCycleTRACER accounts for cell cycle and volume in mass cytometry data. Nat. Commun. 9, 632 (2018).
Kim, T. H. et al. Broadly permissive intestinal chromatin underlies lateral inhibition and cell plasticity. Nature 506, 511–515 (2014).
Levine, J. H. et al. Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell 162, 184–197 (2015).
Orlova, D. Y. et al. Earth Mover’s Distance (EMD): a true metric for comparing biomarker expression levels in cell populations. PLoS ONE 11, e0151859 (2016).
Krishnaswamy, S. et al. Systems biology. Conditional density-based analysis of T cell signaling in single-cell data. Science 346, 1250689 (2014).
Nusse, R. & Clevers, H. Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. Cell 169, 985–999 (2017).
Gehart, H. & Clevers, H. Tales from the crypt: new insights into intestinal stem cells. Nat. Rev. Gastroenterol. Hepatol. 16, 19–34 (2019).
Massague, J. TGFβ signalling in context. Nat. Rev. Mol. Cell Biol. 13, 616–630 (2012).
Zunder, E. R. et al. Palladium-based mass tag cell barcoding with a doublet-filtering scheme and single-cell deconvolution algorithm. Nat. Protoc. 10, 316–333 (2015).
Bodenmiller, B. et al. Multiplexed mass cytometry profiling of cellular states perturbed by small-molecule regulators. Nat. Biotechnol. 30, 858–867 (2012).
Willis, L. M. et al. Tellurium-based mass cytometry barcode for live and fixed cells. Cytom. A 93, 685–694 (2018).
McCarthy, R. L., Mak, D. H., Burks, J. K. & Barton, M. C. Rapid monoisotopic cisplatin based barcoding for multiplexed mass cytometry. Sci. Rep. 7, 3779 (2017).
The Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337 (2012).
Isella, C. et al. Stromal contribution to the colorectal cancer transcriptome. Nat. Genet. 47, 312–319 (2015).
Calon, A. et al. Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat. Genet. 47, 320–329 (2015).
Lan, J. et al. M2 macrophage-derived exosomes promote cell migration and invasion in colon cancer. Cancer Res. 79, 145–158 (2018).
Tape, C. J. The heterocellular emergence of colorectal cancer. Trends Cancer 3, 79–88 (2017).
Dow, L. E. et al. Apc restoration promotes cellular differentiation and reestablishes crypt homeostasis in colorectal cancer. Cell 161, 1539–1552 (2015).
O’Rourke, K. P. et al. Transplantation of engineered organoids enables rapid generation of metastatic mouse models of colorectal cancer. Nat. Biotechnol. 35, 577–582 (2017).
Spitzer, M. H. et al. IMMUNOLOGY. An interactive reference framework for modeling a dynamic immune system. Science 349, 1259425 (2015).
Cruz-Acuna, R. et al. Synthetic hydrogels for human intestinal organoid generation and colonic wound repair. Nat. Cell Biol. 19, 1326–1335 (2017).
Neal, J. T. et al. Organoid modeling of the tumor immune microenvironment. Cell 175, 1972–1988.e16 (2018).
van de Wetering, M. et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 161, 933–945 (2015).
Vlachogiannis, G. et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 359, 920–926 (2018).
Machado, L. et al. In situ fixation redefines quiescence and early activation of skeletal muscle stem cells. Cell Rep. 21, 1982–1993 (2017).
Han, G., Spitzer, M. H., Bendall, S. C., Fantl, W. J. & Nolan, G. P. Metal-isotope-tagged monoclonal antibodies for high-dimensional mass cytometry. Nat. Protoc. 13, 2121–2148 (2018).
Han, G. et al. Atomic mass tag of bismuth-209 for increasing the immunoassay multiplexing capacity of mass cytometry. Cytom. A 91, 1150–1163 (2017).
Chevrier, S. et al. Compensation of signal spillover in suspension and imaging mass cytometry. Cell Syst. 6, 612–620 e615 (2018).
Finck, R. et al. Normalization of mass cytometry data with bead standards. Cytom. A 83, 483–494 (2013).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Khalil, H., Nie, W., Edwards, R. A. & Yoo, J. Isolation of primary myofibroblasts from mouse and human colon tissue. J. Vis. Exp. https://doi.org/10.3791/50611 (2013).
McInnes, L., Healy, J. & Melville, J. UMAP: Uniform Manifold Approximation and Projection for dimension reduction. Preprint at https://arxiv.org/abs/1802.03426 (2018).
van Dijk, D. et al. Recovering gene interactions from single-cell data using data diffusion. Cell 174, 716–729.e27 (2018).
Acknowledgements
We are grateful to L. Dow (Cornell University) for sharing CRC organoids, X. Lu (University of Oxford) for sharing murine intestines and O. Ornatsky (Fluidigm) for providing monoisotopic cisplatin (195Pt and 196Pt). We thank the UCL CI Flow-Core for MC support and L. McInnes for UMAP advice. Graphical organoid renderings were designed by J. Claus (Phospho Biomedical Animation). This work was supported by Cancer Research UK (grant no. C60693/A23783, C.J.T.), UCLH Biomedical Research Centre (grant no. BRC422, C.J.T.), The Royal Society (grant no. RSG\R1\180234, C.J.T.) and Rosetrees Trust (grant no. A1989, C.J.T.).
Author information
Authors and Affiliations
Contributions
X.Q. designed the study, performed organoid and MC experiments, analyzed the data and wrote the paper. J.S. developed TOBis, designed rare-earth metal antibody panels, performed MC analysis and analyzed data. P.V. isolated and characterized colonic fibroblasts, macrophages and cultured organoids, and analyzed data. P.K. performed UMAP, EMD, DREMI and PCA data analysis. M.N. developed TeMal barcodes. S.E.A. provided murine monocytes and intestines for fibroblast isolation. V.S.W.L. provided murine small intestines for organoid isolation. C.J.T. designed the study, analyzed the data and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
M.N. has pending intellectual property on the use of tellurium reagents for mass cytometry applications which has been licensed to Fluidigm Corporation.
Additional information
Peer review information Nina Vogt and Nicole Rusk were the primary editors on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Rare-Earth Metal-Conjugated Antibodies for Organoid Cell-Type Identification.
a) Summary of reagents used in organoid directed differentiation. b) Fuzzy logic diagram of organoid directed differentiation. c) Confocal immunofluorescence (IF) of small intestinal organoids stained with rare-earth metal-conjugated mass cytometry (MC) antibodies showing individual cell-type markers (red), F-Actin (white), and DAPI (blue) following directed differentiation, scale bars = 50 μm. Images are representative of three independent experiments. d) UMAP (Uniform Manifold Approximation and Projection) distributions of ~20,000 single organoid cells analyzed by MC following directed differentiation. e) Force-directed Scaffold maps of directedly differentiated small intestinal organoids. Cell-type landmark nodes are identified from untreated small intestinal organoids. f) Earth Mover’s Distance (EMD) quantification of cell-type identification markers from MC analysis of Lgr5-EGFP-ires-CreERT2 small intestinal crypt-cells.
Supplementary Figure 2 Organoid Cell-Type and Cell-State Identification.
a) Strategy for organoid cell-type identification. b) Strategy for organoid cell-state classification. c) EMD quantification of cell-type identification markers across all small intestinal organoid cell-types from Figs. 1 and 2. d) Cell-state quantification of all small intestinal organoid cell-types from Figs. 1 and 2.
Supplementary Figure 3 Cell-Type and Cell-State Specific Signaling Networks in Intestinal Organoids.
Cell-type and cell-state specific signaling networks of 27 PTMs from ~1 million single organoid cells analyzed by MC, with nodes colored by PTM-EMD scores quantifying PTM intensity (relative to all organoid cells), and edges colored by DREMI scores quantifying PTM-PTM connectivity.
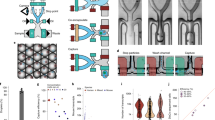
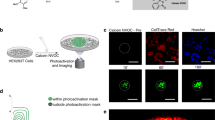
Supplementary Figure 4 Maxpar versus TOBis Organoid Barcoding in situ.
a) De-barcoded cell counts from small intestinal organoids barcoded using the Maxpar Cell-IDTM Barcoding Kit either after removal from Matrigel (ex situ) or while still in Matrigel (in situ) (n = 2 independent samples). b) Mean dual counts of 110Pd from Matrigel incubated in PBS, Maxpar Cell-IDTM barcode #20 (106Pd, 108Pd, and 110Pd) resuspended in PBS (PBS + BC), or Maxpar Cell-IDTM barcode #20 resuspended in Maxpar Cell Staining Buffer (CSB + BC) (two-tailed unpaired t-test, p < 0.0001, n = 3 independent samples). Error bars represent standard deviation (SD). c) Maximum-gain confocal IF images of NHS ester-/C2 maleimide-Alexa 647 fluorescent probes shown in Fig. 3a to highlight background Matrigel staining. Each IF image is representative of three independent experiments. d) Small intestinal organoids concurrently stained in situ with Maxpar Cell-IDTM (102Pd, 104Pd, 105Pd, 106Pd, 108Pd, and 110Pd) and TOBis (124Te, 126Te, 128Te, 130Te, 196Pt, and 198Pt) barcodes using analogous 20-plex 6-choose-3 matrices. Data is representative of three independent experiments.
Supplementary Figure 5 Maxpar versus TOBis Cell-Recovery Comparison.
a) Example 10-plex barcoding workflow for organoid MC using Maxpar Cell-IDTM Barcoding Kit. b) Example 10-plex barcoding workflow for organoid MC using TOBis. c) Dissociation and d) centrifugation steps (s) required for multiplexed organoid experiments using Maxpar or TOBis barcoding. e) Single-cell recovery of serially-titrated small intestinal organoids barcoded with either Maxpar (ex situ) or TOBis (in situ) (two-tailed ratio-paired t-test, p < 0.001, n = 2 independent set of serially-titrated samples). f) Cell-type recovery (cell count) from equally seeded small intestinal organoids analyzed by MC and de-barcoded by Maxpar or TOBis. g) Cell-type recovery (population percentage) from equally sampled small intestinal organoid cells analyzed by MC and de-barcoded by Maxpar or TOBis (linear regression of correlation, R2 = 0.86, n = 2 independent samples). h) Mean debarcoded cell counts in the absence or presence of glutathione (GSH) in PBS washes following TOBis barcoding (two-tailed unpaired t-test, p < 0.0001, n = 3 independent samples). Error bars represent SD..
Supplementary Figure 6 Colorectal Cancer (CRC) Tumor Microenvironment (TME) Model Cells.
a) Confocal IF of CRC organoid genotypes (wild-type (WT), shApc (A), shApc and KrasG12D/+ (AK), and shApc, KrasG12D/+ and Trp53R172H/- (AKP)) stained with Pan-CK, LRIG1, CD44, and C-MYC (red), F-Actin (white), and DAPI (blue), scale bars = 50 μm. Images are representative of three independent IF experiments. Pan-CK and LRIG1 (in combination with CEACAM1) were subsequently used for epithelial and stem cell identification in MC. b) Confocal IF of colonic fibroblasts cultured in 2D (plastic) and 3D (Matrigel) demonstrates positive staining for colonic fibroblast markers PDPN, Vimentin, PDGFRɑ, FOXL1, and GLI1 (red), F-Actin (white), and DAPI (blue), scale bars = 50 μm. Images are representative of three independent IF experiments. PDPN and RFP were subsequently used for fibroblast identification in MC. c) Confocal IF of primary bone marrow derived macrophages cultured in 2D (plastic) and 3D (Matrigel) demonstrates positive staining for macrophages markers CD68, F4/80, CD45, CD11b, and CX3CR1 (red), F-Actin (white), and DAPI (blue), scale bars = 50 μm. Images are representative of three independent IF experiments. CD68 and F40/80 were subsequently used for macrophage identification in MC.
Supplementary Figure 7 CRC Organoid Signaling Networks in the CRC TME Model.
Signaling networks of 28 PTMs in colonic organoid cells from each genotype (wild-type (WT), shApc (A), shApc and KrasG12D/+ (AK), or shApc, KrasG12D/+, and Trp53R172H/- (AKP)) and microenvironment condition in the CRC TME model analyzed by MC. Signaling nodes are connected by DREMI edges and PTM activity is displayed as EMD relative to all cells across all conditions.
Supplementary Figure 8 Macrophage and Fibroblast Signaling Profiles in the CRC TME Model.
a) and b) Signaling networks of 28 PTMs in macrophages and colonic fibroblasts from each genotype and microenvironment condition in the CRC TME model analyzed by MC. Signaling nodes are connected by DREMI edges and PTM activity is displayed as EMD relative to all cells across all conditions. c) and d) PCA of 28 PTM-EMDs for macrophages and colonic fibroblasts across all genotype/microenvironment combinations (10 conditions each). e) and f) PCA of 756 PTM-DREMIs for macrophages and colonic fibroblasts across all genotype/microenvironment combinations (10 conditions each).
Supplementary Figure 9 CRC Organoid Microenvironment and Genotype Scaffold Maps.
a) Force-directed Scaffold maps were constructed from WT colonic organoids alone or co-cultured with macrophages and/or colonic fibroblasts. b) Force-directed Scaffold maps were constructed from WT, A, AK, and AKP colonic organoid monocultures.
Supplementary Information
Supplementary Information
Supplementary Figs. 1–9 and Tables 1–4.
Supplementary Table 5
Parameters for single-cell signaling data analysis markers selected and parameters used for UMAP, PCA and Scaffold analysis.
Rights and permissions
About this article
Cite this article
Qin, X., Sufi, J., Vlckova, P. et al. Cell-type-specific signaling networks in heterocellular organoids. Nat Methods 17, 335–342 (2020). https://doi.org/10.1038/s41592-020-0737-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-020-0737-8
This article is cited by
-
Establishment of gastrointestinal assembloids to study the interplay between epithelial crypts and their mesenchymal niche
Nature Communications (2023)
-
Intestinal cellular heterogeneity and disease development revealed by single-cell technology
Cell Regeneration (2022)
-
Traject3d allows label-free identification of distinct co-occurring phenotypes within 3D culture by live imaging
Nature Communications (2022)
-
The promise of organoids for unraveling the proteomic landscape of the developing human brain
Molecular Psychiatry (2022)
-
A microenvironment-inspired synthetic three-dimensional model for pancreatic ductal adenocarcinoma organoids
Nature Materials (2022)