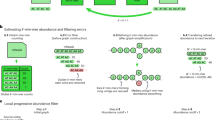
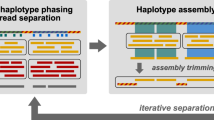
Abstract
Long-read sequencing technologies have substantially improved the assemblies of many isolate bacterial genomes as compared to fragmented short-read assemblies. However, assembling complex metagenomic datasets remains difficult even for state-of-the-art long-read assemblers. Here we present metaFlye, which addresses important long-read metagenomic assembly challenges, such as uneven bacterial composition and intra-species heterogeneity. First, we benchmarked metaFlye using simulated and mock bacterial communities and show that it consistently produces assemblies with better completeness and contiguity than state-of-the-art long-read assemblers. Second, we performed long-read sequencing of the sheep microbiome and applied metaFlye to reconstruct 63 complete or nearly complete bacterial genomes within single contigs. Finally, we show that long-read assembly of human microbiomes enables the discovery of full-length biosynthetic gene clusters that encode biomedically important natural products.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Sequencing data for the sheep gut sample are available under the NCBI BioProject PRJNA595610. HMP mock dataset is available at: https://github.com/PacificBiosciences/DevNet/wiki/Human_Microbiome_Project_MockB_Shotgun. Zymo datasets are at: https://github.com/LomanLab/mockcommunity. Cow rumen dataset is at: NCBI SRA repository under BioProject PRJNA507739. Human stool samples are at: ENA project PRJEB29152. NCBI accession codes for the sequences used in the NRPS analysis are: AM229678.1, AB101202.1, FP929054.1 and FP929054.1. All assemblies that were evaluated in this study, as well as SYNTH64 and SYNTH181 datasets are available at: https://doi.org/10.5281/zenodo.3986210 (ref. 66).
Code availability
metaFlye is freely available as a part of the Flye package at: https://github.com/fenderglass/Flye. The pbclip tool for PacBio subread splitting is available from https://github.com/fenderglass/pbclip.
References
Jain, M. et al. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol. 36, 338 (2018).
Miga, K. H. et al. Telomere-to-telomere assembly of a complete human X chromosome. Nature https://doi.org/10.1038/s41586-020-2547-7 (2020).
Tsai, Y. C. et al. Resolving the complexity of human skin metagenomes using single-molecule sequencing. MBio 7, e01948–15 (2016).
Driscoll, C. B., Otten, T. G., Brown, N. M. & Dreher, T. W. Towards long-read metagenomics: complete assembly of three novel genomes from bacteria dependent on a diazotrophic cyanobacterium in a freshwater lake co-culture. Stand. Genom. Sci. 12, 9 (2017).
Nicholls, S. M., Quick, J. C., Tang, S. & Loman, N. J. Ultra-deep, long-read nanopore sequencing of mock microbial community standards. GigaScience 8, 1–9 (2019).
Bertrand, D. et al. Hybrid metagenomic assembly enables high-resolution analysis of resistance determinants and mobile elements in human microbiomes. Nat. Biotechnol. 37, 937–944 (2019).
Somerville, V. et al. Long read-based de novo assembly of low complex metagenome samples results in finished genomes and reveals insights into strain diversity and an active phage system. BMC Microbiol. 19, 143 (2019).
Moss, E. L., Maghini, D. G. & Bhatt, A. S. Complete, closed bacterial genomes from microbiomes using nanopore sequencing. Nat. Biotechnol. 38, 701–707 (2020).
Stewart, R. D. et al. Compendium of 4,941 rumen metagenome-assembled genomes for rumen microbiome biology and enzyme discovery. Nat. Biotechnol. 37, 953–961 (2019).
Arumugam, K. et al. Annotated bacterial chromosomes from frame-shift-corrected long read metagenomic data. Microbiome 7, 61 (2019).
Hiraoka, S. et al. Metaepigenomic analysis reveals the unexplored diversity of DNA methylation in an environmental prokaryotic community. Nat. Commun. 10, 159 (2019).
Bickhart, D. M. et al. Assignment of virus and antimicrobial resistance genes to microbial hosts in a complex microbial community by combined long-read assembly and proximity ligation. Genome Biol. 20, 1–18 (2019).
Chin, C. S. et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods 13, 1050–1054 (2016).
Li, H. Minimap and miniasm: fast mapping and de novo assembly for noisy long sequences. Bioinformatics 32, 2103–2110 (2016).
Koren, S. et al. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736 (2017).
Kolmogorov, M., Yuan, J., Lin, Y. & Pevzner, P. A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 37, 540–546 (2019).
Ruan, J. & Li, H. Fast and accurate long-read assembly with wtdbg2. Nat. Methods 17, 155–158 (2020).
Li, D., Liu, C. M., Luo, R., Sadakane, K. & Lam, T. W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674–1676 (2015).
Nurk, S., Meleshko, D., Korobeynikov, A. & Pevzner, P. A. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 27, 824–834 (2017).
Truong, D. T., Tett, A., Pasolli, E., Huttenhower, C. & Segata, N. Microbial strain-level population structure and genetic diversity from metagenomes. Genome Res. 27, 626–638 (2017).
Ghurye, J., Treangen, T., Fedarko, M., Hervey, W. J. & Pop, M. MetaCarvel: linking assembly graph motifs to biological variants. Genome Biol. 20, 174 (2019).
Goltsman, D. S. A. et al. Metagenomic analysis with strain-level resolution reveals fine-scale variation in the human pregnancy microbiome. Genome Res. 28, 1467–1480 (2018).
Guo, J. et al. Horizontal gene transfer in an acid mine drainage microbial community. BMC Genomics 16, 496 (2015).
Eloe-Fadrosh, E. A. et al. Global metagenomic survey reveals a new bacterial candidate phylum in geothermal springs. Nat. Commun. 7, 10476 (2016).
Suzuki, Y. et al. Long-read metagenomic exploration of extrachromosomal mobile genetic elements in the human gut. Microbiome 7, 119 (2019).
Stevenson, L. J., Owen, J. G. & Ackerley, D. F. Metagenome driven discovery of nonribosomal peptides. ACS Chem. Biol. 14, 2115–2126 (2019).
Nijkamp, J. F., Pop, M., Reinders, M. J. T. & de Ridder, D. Exploring variation-aware contig graphs for (comparative) metagenomics using MaryGold. Bioinformatics 29, 2826–2834 (2013).
Onodera, T., Sadakane, K. & Shibuya, T. Detecting superbubbles in assembly graphs. In International Workshop on Algorithms in Bioinformatics, 338–348 (Springer, 2013).
Garg, S. et al. A haplotype-aware de novo assembly of related individuals using pedigree sequence graph. Bioinformatics 36, 2385–2392 (2020).
Sczyrba, A. et al. Critical assessment of metagenome interpretation - a benchmark of metagenomics software. Nat. Methods 14, 1063–1071 (2017).
Jain, C., Rodriguez-R, L. M., Phillippy, A. M., Konstantinidis, K. T. & Aluru, S. High throughput ANI analysis of 90 K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9, 1–8 (2018).
Wick, R. Badread: simulation of error-prone long reads. J. Open Source Softw. 4, 1316 (2019).
Mikheenko, A., Prjibelski, A., Saveliev, V., Antipov, D. & Gurevich, A. Versatile genome assembly evaluation with QUAST-LG. Bioinformatics 34, i142–i150 (2018).
Vaser, R., Sović, I., Nagarajan, N. & Šikić, M. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 27, 737–746 (2017).
Antipov, D., Raiko, M., Lapidus, A. & Pevzner, P. A. Plasmid detection and assembly in genomic and metagenomic data sets. Genome Res. 29, 961–968 (2019).
Latorre-Pérez, Adriel, Villalba-Bermell, Pascual, Pascual, Javier & Vilanova, Cristina Assembly methods for nanopore-based metagenomic sequencing: a comparative study. Sci. Rep. 10, 1–14 (2020).
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P. & Tyson, G. W. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055 (2015).
Hyatt, D. et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinf. 11, 119 (2010).
Laetsch, D. R. & Blaxter, M. L. BlobTools: interrogation of genome assemblies. F1000Research 6, 1287 (2017).
Buchfink, B., Xie, C. & Huson, D. H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60 (2015).
UniProt Consortium. UniProt: a hub for protein information. Nucleic Acids Res. 43, D204–D212 (2014).
Walker, B. J. et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 9, e112963 (2014).
Minkin, I. & Medvedev, P. Scalable multiple whole-genome alignment and locally collinear block construction with SibeliaZ. Preprint at bioRxiv https://doi.org/10.1101/548123 (2019).
Kersten, R. D. et al. A mass spectrometry-guided genome mining approach for natural product peptidogenomics. Nat. Chem. Biol. 7, 794–802 (2011).
Ling, L. L. et al. A new antibiotic kills pathogens without detectable resistance. Nature 517, 455–459 (2015).
Meleshko, D. et al. BiosyntheticSPAdes: reconstructing biosynthetic gene clusters from assembly graphs. Genome Res. 29, 1352–1362 (2019).
Behsaz, B. et al. De novo peptide sequencing reveals many cyclopeptides in the human gut and other environments. Cell Syst. 10, 99–108 (2020).
Wilson, M. R. et al. The human gut bacterial genotoxin colibactin alkylates DNA. Science 363, eaar7785 (2019).
Mohimani, H. & Pevzner, P. A. Dereplication, sequencing and identification of peptidic natural products: from genome mining to peptidogenomics to spectral networks. Nat. Prod. Rep. 33, 73–86 (2016).
Quast, C. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596 (2012).
Wick, R. R., Schultz, M. B., Zobel, J. & Holt, K. E. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 31, 3350–3352 (2015).
Hunter, J. D. Matplotlib A 2D graphics environment. Comput. Sci. Eng. 9, 90 (2007).
Dolev, S., Ghanayim, M., Binun, B., Frenkel, S. & Sun, Y. S. Relationship of Jaccard and edit distance in malware clustering and online identification. In 2017 IEEE 16th International Symposium on Network Computing and Applications (NCA), 1–5 (IEEE, 2017).
Fu, L., Niu, B., Zhu, Z., Wu, S. & Li, W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152 (2012).
Rognes, T., Flouri, T., Nichols, B., Quince, C. & Mahé, F. VSEARCH: a versatile open source tool for metagenomics. PeerJ 4, e2584 (2016).
Pruitt, K. D., Tatusova, T. & Maglott, D. R. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 35, D61–D65 (2007).
Li, X., Andersen, D. G., Kaminsky, M. & Freedman, M. J. Algorithmic improvements for fast concurrent cuckoo hashing. In Proceedings of the Ninth European Conference on Computer Systems, 27 (ACM, 2014).
Jiang, Z. et al. Ancestral reconstruction of segmental duplications reveals punctuated cores of human genome evolution. Nat. Genet. 39, 1361–1368 (2007).
Bankevich, A. & Pevzner, P. A. mosaicFlye: resolving long mosaic repeats using long error-prone reads. Preprint at bioRxiv, https://doi.org/10.1101/2020.01.15.908285 (2020).
Koren, S., Treangen, T. J. & Pop, M. Bambus 2: scaffolding metagenomes. Bioinformatics 27, 2964–2971 (2011).
Li, H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018).
Nurk, S. et al. Assembling genomes and mini-metagenomes from highly chimeric reads. J. Comp. Biol. 20, 714–737 (2013).
Brankovic, L. et al. Linear-time superbubble identification algorithm for genome assembly. Theor. Comput. Sci. 609, 374–383 (2016).
Zerbino, D. R. & Birney, E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18, 821–829 (2008).
Paten, B. et al. Superbubbles, ultrabubbles, and cacti. J. Computational Biol. 25, 649–663 (2018).
Supporting data for the manuscript “metaFlye: scalable long-read metagenome assembly using repeat graphs” (version 3.0) (Dataset). Zenodo https://doi.org/10.5281/zenodo.3986210 (2020).
Acknowledgements
We are grateful to Denis Bertrand and Niranjan Nagarajan for sharing the metagenomic datasets before journal publication. M.K. and P.A.P. were supported by the NSF/MCB-BSF grant 1715911. B.B. was supported by the US National Institutes of Health grant 2-P41-GM103484. D.B. was funded by USDA CRIS project 5090-31000-026-00-D and K.K., S.S. and T.S. by project 3040-31000-100-00D. A.G. and M.R. were supported by the Russian Science Foundation (grant 19-16-00049). Computational resources were provided in part by the Research Park Computer Center at St. Petersburg State University.
Author information
Authors and Affiliations
Contributions
M.K., J.Y. and P.P. developed the metaFlye concept. M.K. implemented and maintained metaFlye. E.P. implemented the short plasmid analysis module. D.B., S.B.S., K.K. and T.S. performed sheep gut sequencing. M.K., D.B., A.G. and M.R. benchmarked metaFlye and analyzed results. A.G. and M.K. performed analysis of synthetic datasets. M.R. analyzed plasmid and virus content. B.B. performed analysis of biosynthetic gene clusters. M.K., D.B., B.B., A.G., M.R., T.S. and P.P. edited the manuscript. P.P. supervised the project. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Lei Tang was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Information about metaFlye, Flye, Canu, miniasm, and wtdbg2 assemblies of the individual genomes in the SYNTH64 dataset.
NGA50 (in megabases) and reference coverage (in percentages) reported for all genomes from the SYNTH64 dataset. Genomes are ordered in the increasing mean NGA50 across all assemblers. Challenging genomes that have closely related species or strains in the metagenome are marked with (!). Grey bars on the NGA50 plot represent the length of the longest chromosome in the reference sequence for each genome (a theoretical upper bound for NGA50). NGA50 is shown in logarithmic scale (not shown for values lower than 100 kb or if the reference coverage is below 50%). The full metaQUAST report for the SYNTH64 dataset is provided in Supplementary Table 1.
Extended Data Fig. 2 NGAx plots for the mock community datasets (HMP mock, ZymoEven GridION, ZymoLog GridION).
NGA(x) is the statistic computed for contigs that are broken at their misassembly breakpoints (if any). NGA(x) is the highest possible number L such that all broken contigs that are longer than L cover at least X% of the reference. Plots were generated by metaQUAST using all available references for each dataset. Flye failed to assemble the ZymoLog datasets due to poor k-mer indexing (Methods).
Extended Data Fig. 3 Base-pair accuracy analysis for assemblies of the mock community datasets (HMP, ZymoEven GridION, and ZymoLog GridION).
Heatmaps showing the number of mismatches and short indels per 100 kbp for each species reference, computed using metaQUAST. Blue and red colors correspond to the values higher and lower than the median, respectively. Statistics were not computed for genomes with no assembled sequence (“-” symbol). Flye failed to assemble the ZymoLog datasets due to poor k-mer indexing (Methods).
Extended Data Fig. 4 The ORF lengths distribution and the GC content distribution of metaFlye and Canu assemblies of the sheep microbiome.
The ORF length distribution suggests similar base-level accuracy for both assemblies.
Extended Data Fig. 5 Taxonomic assignments of sheep microbiome assemblies.
a, metaFlye contigs assignment at the phylum level visualized with BlobTools. b, Length distributions of metaFlye and Canu contigs within each assigned superkingdom.
Extended Data Fig. 6 Statistics of simple bubbles for the metaFlye assemblies human gut and cow rumen.
(Left) the human gut dataset with 615 bubbles, and (right) the cow rumen dataset with 1510 bubbles. Bubble counts exclude loops, and include roundabouts with two edges.
Extended Data Fig. 7 Analysis of sequence overlap between 19 human gut samples.
Multi-way sequence alignments were computed using SiebliaZ. (left) The proportions of unique and shared sequences in each sample. An assembled segment within a sample is called unique if it has no alignments against sequence from any other samples. Otherwise, the segment is shared. (right) The total amount of sequence for each multiplicity bin. A sequence fragment belongs to the multiplicity bin X if it is shared by exactly X samples.
Supplementary information
Supplementary Information
Supplementary Tables 4–10 and Notes 1–8
Supplementary Table 1
Detailed information about metaFlye, Flye, Canu, miniasm and wtdbg2 assemblies of the SYNTH64 dataset
Supplementary Table 2
Detailed information about metaFlye, Flye, Canu, miniasm and wtdbg2 assemblies of the SYNTH181 dataset
Supplementary Table 3
Detailed information about metaFlye, Flye, Canu, miniasm and wtdbg2 assemblies of all mock community datasets
Rights and permissions
About this article
Cite this article
Kolmogorov, M., Bickhart, D.M., Behsaz, B. et al. metaFlye: scalable long-read metagenome assembly using repeat graphs. Nat Methods 17, 1103–1110 (2020). https://doi.org/10.1038/s41592-020-00971-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-020-00971-x
This article is cited by
-
MetageNN: a memory-efficient neural network taxonomic classifier robust to sequencing errors and missing genomes
BMC Bioinformatics (2024)
-
Evaluating and improving the representation of bacterial contents in long-read metagenome assemblies
Genome Biology (2024)
-
Novel functional insights into the microbiome inhabiting marine plastic debris: critical considerations to counteract the challenges of thin biofilms using multi-omics and comparative metaproteomics
Microbiome (2024)
-
Recovering high-quality bacterial genomes from cross-contaminated cultures: a case study of marine Vibrio campbellii
BMC Genomics (2024)
-
Unraveling metagenomics through long-read sequencing: a comprehensive review
Journal of Translational Medicine (2024)