Abstract
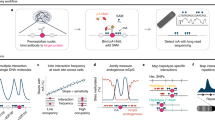
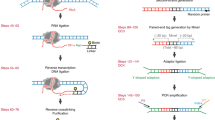
Mapping open chromatin regions has emerged as a widely used tool for identifying active regulatory elements in eukaryotes. However, existing approaches, limited by reliance on DNA fragmentation and short-read sequencing, cannot provide information about large-scale chromatin states or reveal coordination between the states of distal regulatory elements. We have developed a method for profiling the accessibility of individual chromatin fibers, a single-molecule long-read accessible chromatin mapping sequencing assay (SMAC-seq), enabling the simultaneous, high-resolution, single-molecule assessment of chromatin states at multikilobase length scales. Our strategy is based on combining the preferential methylation of open chromatin regions by DNA methyltransferases with low sequence specificity, in this case EcoGII, an N6-methyladenosine (m6A) methyltransferase, and the ability of nanopore sequencing to directly read DNA modifications. We demonstrate that aggregate SMAC-seq signals match bulk-level accessibility measurements, observe single-molecule nucleosome and transcription factor protection footprints, and quantify the correlation between chromatin states of distal genomic elements.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Short-read datasets associated with this study are available through GEO accession GSE128290. Nanopore data are available through SRA BioProject PRJNA594057. Nanopore raw data are available at https://zoharshiponh.s3.amazonaws.com/NMETH_2020/index.html.
References
Wu, C. The 5′ ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature 286, 854–860 (1980).
Keene, M. A., Corces, V., Lowenhaupt, K. & Elgin, S. C. DNase I hypersensitive sites in Drosophila chromatin occur at the 5′ ends of regions of transcription. Proc. Natl Acad. Sci. USA 78, 143–146 (1981).
McGhee, J. D., Wood, W. I., Dolan, M., Engel, J. D. & Felsenfeld, G. A 200 base pair region at the 5′ end of the chicken adult β-globin gene is accessible to nuclease digestion. Cell 27, 45–55 (1981).
Dorschner, M. O. et al. High-throughput localization of functional elements by quantitative chromatin profiling. Nat. Methods 1, 219–225 (2004).
Sabo, P. J. et al. Genome-scale mapping of DNase I sensitivity in vivo using tiling DNA microarrays. Nat. Methods 3, 511–518 (2006).
Boyle, A. P. et al. High-resolution mapping and characterization of open chromatin across the genome. Cell 132, 311–322 (2008).
Hesselberth, J. R. et al. Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat. Methods 6, 283–289 (2009).
Schones, D. E. et al. Dynamic regulation of nucleosome positioning in the human genome. Cell 132, 887–898 (2008).
Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y. & Greenleaf, W. J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013).
Kelly, T. K. et al. Genome-wide mapping of nucleosome positioning and DNA methylation within individual DNA molecules. Genome Res. 22, 2497–2506 (2012).
Nabilsi, N. H. et al. Multiplex mapping of chromatin accessibility and DNA methylation within targeted single molecules identifies epigenetic heterogeneity in neural stem cells and glioblastoma. Genome Res. 24, 329–339 (2014).
Krebs, A. R. et al. Genome-wide single-molecule footprinting reveals high RNA polymerase II turnover at paused promoters. Mol. Cell 67, 411–422.e4 (2017).
Murray, I. A. et al. The non-specific adenine DNA methyltransferase M.EcoGII. Nucleic Acids Res. 46, 840–848 (2018).
Simpson, J. T. et al. Detecting DNA cytosine methylation using nanopore sequencing. Nat. Methods 14, 407–410 (2017).
Rand, A. C. et al. Mapping DNA methylation with high-throughput nanopore sequencing. Nat. Methods 14, 411–413 (2017).
Feng, S. et al. Conservation and divergence of methylation patterning in plants and animals. Proc. Natl Acad. Sci. USA 107, 8689–8694 (2010).
Zemach, A., McDaniel, I. E., Silva, P. & Zilberman, D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 328, 916–919 (2010).
Lister, R. et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133, 523–536 (2008).
Brogaard, K., Xi, L., Wang, J. P. & Widom, J. A map of nucleosome positions in yeast at base-pair resolution. Nature 486, 496–501 (2012).
Stoiber, M. H. et al. De novo identification of DNA modifications enabled by genome-guided nanopore signal processing. Preprint at bioRxiv https://doi.org/10.1101/094672 (2017).
Li, H. Minimap and miniasm: fast mapping and de novo assembly for noisy long sequences. Bioinformatics 32, 2103–2110 (2016).
Cole, H. A., Howard, B. H. & Clark, D. J. The centromeric nucleosome of budding yeast is perfectly positioned and covers the entire centromere. Proc. Natl Acad. Sci. USA 108, 12687–12692 (2011).
Henikoff, S. et al. The budding yeast centromere DNA element II wraps a stable Cse4 hemisome in either orientation in vivo. Elife 3, e01861 (2014).
Conconi, A., Widmer, R. M., Koller, T. & Sogo, J. M. Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell 57, 753–761 (1989).
French, S. L., Osheim, Y. N., Cioci, F., Nomura, M. & Beyer, A. L. In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Mol. Cell Biol. 23, 1558–1568 (2003).
Goetze, H. et al. Alternative chromatin structures of the 35S rRNA genes in Saccharomyces cerevisiae provide a molecular basis for the selective recruitment of RNA polymerases I and II. Mol. Cell Biol. 30, 2028–2045 (2010).
Merz, K. et al. Actively transcribed rRNA genes in S. cerevisiae are organized in a specialized chromatin associated with the high-mobility group protein Hmo1 and are largely devoid of histone molecules. Genes Dev. 22, 1190–1204 (2008).
Panday, A. & Grove, A. Yeast HMO1: linker histone reinvented. Microbiol. Mol. Biol. Rev. 81, e00037–16 (2016).
Jones, H. S. et al. RNA polymerase I in yeast transcribes dynamic nucleosomal rDNA. Nat. Struct. Mol. Biol. 14, 123–130 (2007).
Huang, J. & Moazed, D. Association of the RENT complex with nontranscribed and coding regions of rDNA and a regional requirement for the replication fork block protein Fob1 in rDNA silencing. Genes Dev. 17, 2162–2176 (2003).
Zhu, F. et al. The interaction landscape between transcription factors and the nucleosome. Nature 562, 76–81 (2018).
O’Sullivan, J. M. et al. Gene loops juxtapose promoters and terminators in yeast. Nat Genet 36, 1014–1018 (2004).
Tan-Wong, S. M. et al. Gene loops enhance transcriptional directionality. Science 338, 671–675 (2012).
Hsieh, T. H. et al. Mapping nucleosome resolution chromosome folding in yeast by Micro-C. Cell 162, 108–119 (2015).
Weiner, A. et al. High-resolution chromatin dynamics during a yeast stress response. Mol. Cell 58, 371–386 (2015).
Morano, K. A., Grant, C. M. & Moye-Rowley, W. S. The response to heat shock and oxidative stress in Saccharomyces cerevisiae. Genetics 190, 1157–1195 (2012).
Gabrieli, T. et al. Selective nanopore sequencing of human BRCA1 by Cas9-assisted targeting of chromosome segments (CATCH). Nucleic Acids Res. 46, e87 (2018).
Staševskij, Z., Gibas, P., Gordevičius, J., Kriukienė, E. & Klimašauskas, S. Tethered oligonucleotide-primed sequencing, TOP-Seq: a high-resolution economical approach for DNA epigenome profiling. Mol. Cell 65, 554–564.e6 (2017).
Fu, Y., Sinha, M., Peterson, C. L. & Weng, Z. The insulator binding protein CTCF positions 20 nucleosomes around its binding sites across the human genome. PLoS Genet. 4, e1000138 (2008).
Fu, Y. et al. N6-methyldeoxyadenosine marks active transcription start sites in Chlamydomonas. Cell 161, 879–892 (2015).
Wang, Y., Chen, X., Sheng, Y., Liu, Y. & Gao, S. N6-adenine DNA methylation is associated with the linker DNA of H2A.Z-containing well-positioned nucleosomes in Pol II-transcribed genes in Tetrahymena. Nucleic Acids Res. 45, 11594–11606 (2017).
Luo, G. Z. et al. N6-methyldeoxyadenosine directs nucleosome positioning in Tetrahymena DNA. Genome Biol. 19, 200 (2018).
Timinskas, A., Butkus, V. & Janulaitis, A. Sequence motifs characteristic for DNA [cytosine-N4] and DNA [adenine-N6] methyltransferases. Classification of all DNA methyltransferases. Gene 157, 3–11 (1995).
Salter, J. D., Bennett, R. P. & Smith, H. C. The APOBEC protein family: united by structure, divergent in function. Trends Biochem. Sci. 41, 578–594 (2016).
Kawasaki, F. et al. Genome-wide mapping of 5-hydroxymethyluracil in the eukaryote parasite Leishmania. Genome Biol. 18, 23 (2017).
Schep, A. N. et al. Structured nucleosome fingerprints enable high-resolution mapping of chromatin architecture within regulatory regions. Genome Res. 25, 1757–1770 (2015).
Miura, F., Enomoto, Y., Dairiki, R. & Ito, T. Amplification-free whole-genome bisulfite sequencing by post-bisulfite adaptor tagging. Nucleic Acids Res. 40, e136 (2012).
Krueger, F. & Andrews, S. R. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571–1572 (2011).
Corces, M. R. et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 14, 959–962 (2017).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Hu, B. et al. Biological chromodynamics: a general method for measuring protein occupancy across the genome by calibrating ChIP-seq. Nucleic Acids Res. 43, e132 (2015).
Feng, J., Liu, T., Qin, B., Zhang, Y. & Liu, X. S. Identifying ChIP-seq enrichment using MACS. Nat. Protoc. 7, 1728–1740 (2012).
Trapnell, C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012).
Grant, C. E., Bailey, T. L. & Noble, W. S. FIMO: scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018 (2011).
Bailey, T. L. et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–208 (2009).
Weirauch, M. T. et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell 158, 1431–1443 (2014).
Pelechano, V., Wei, W. & Steinmetz, L. M. Extensive transcriptional heterogeneity revealed by isoform profiling. Nature 497, 127–131 (2013).
Ramachandran, S., Zentner, G. E. & Henikoff, S. Asymmetric nucleosomes flank promoters in the budding yeast genome. Genome Res. 25, 381–390 (2015).
Chereji, R. V., Ramachandran, S., Bryson, T. D. & Henikoff, S. Precise genome-wide mapping of single nucleosomes and linkers in vivo. Genome Biol. 19, 19 (2018).
Acknowledgements
This work was supported by National Institutes of Health grants (nos. P50HG007735, RO1 HG008140, U19AI057266 and UM1HG009442 to W.J.G., 1UM1HG009436 to W.J.G. and A.K., 1DP2OD022870-01 and 1U01HG009431 to A.K.), the Rita Allen Foundation (to W.J.G.), the Baxter Foundation Faculty Scholar Grant and the Human Frontiers Science Program grant RGY006S (to W.J.G). W.J.G is a Chan Zuckerberg Biohub investigator and acknowledges grant nos. 2017-174468 and 2018-182817 from the Chan Zuckerberg Initiative. Z.S. is supported by EMBO Long-Term Fellowship EMBO ALTF 1119-2016 and by Human Frontier Science Program Long-Term Fellowship HFSP LT 000835/2017-L. G.K.M. is supported by the Stanford School of Medicine Dean’s Fellowship. N.A.S.A. is funded by the Department of Defense through a National Defense Science and Engineering Grant and by a Stanford Graduate Fellowship. We also thank members of the Greenleaf and Kundaje laboratories for their helpful suggestions and discussions on the subject over the course of the study.
Author information
Authors and Affiliations
Contributions
Z.S., G.K.M. and N.A.S.A. conceived and designed the study. Z.S., G.K.M. and N.A.S.A. performed initial experiments. Z.S., M.P.S. and G.K.M. performed diamide time course experiments. G.K.M. and Z.S. analyzed data. W.J.G., A.K. and J.M.S. supervised the study. G.K.M., Z.S., W.J.G. and A.K. wrote the manuscript with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nicole Rusk and Lei Tang were the primary editors on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Tables 1–4 and Figs. 1–73.
Rights and permissions
About this article
Cite this article
Shipony, Z., Marinov, G.K., Swaffer, M.P. et al. Long-range single-molecule mapping of chromatin accessibility in eukaryotes. Nat Methods 17, 319–327 (2020). https://doi.org/10.1038/s41592-019-0730-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-019-0730-2
This article is cited by
-
Multimodal epigenetic sequencing analysis (MESA) of cell-free DNA for non-invasive colorectal cancer detection
Genome Medicine (2024)
-
Single-nucleoid architecture reveals heterogeneous packaging of mitochondrial DNA
Nature Structural & Molecular Biology (2024)
-
NOMe-HiC: joint profiling of genetic variant, DNA methylation, chromatin accessibility, and 3D genome in the same DNA molecule
Genome Biology (2023)
-
BIND&MODIFY: a long-range method for single-molecule mapping of chromatin modifications in eukaryotes
Genome Biology (2023)
-
Single-cell sequencing technology applied to epigenetics for the study of tumor heterogeneity
Clinical Epigenetics (2023)