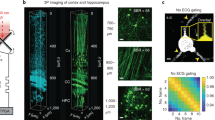
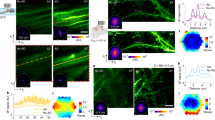
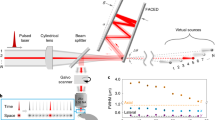
Abstract
Optical imaging is important for understanding brain function. However, established methods with high spatiotemporal resolution are limited by the potential for laser damage to living tissues. We describe an adaptive femtosecond excitation source that only illuminates the region of interest, which leads to a 30-fold reduction in the power requirement for two- or three-photon imaging of brain activity in awake mice for improved high-speed longitudinal neuroimaging.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Code availability
The MATLAB code for driving the AWG (NI PXI-5412, National Instruments) and data-acquisition card (ATS9371, Alazar Technologies) can be found at https://github.com/Bo-Li-ORCID-0000-0003-1492-0919/Adaptive-excitation-source-for-high-speed-multi-photon-microscopy.
References
Denk, W., Strickler, J. H. & Webb, W. W. Two-photon laser scanning fluorescence microscopy. Science 248, 73–76 (1990).
Zipfel, W. R., Williams, R. M. & Webb, W. W. Nonlinear magic: multiphoton microscopy in the biosciences. Nat. Biotechnol. 21, 1369–1377 (2003).
Helmchen, F. & Denk, W. Deep tissue two-photon microscopy. Nat. Meth. 2, 932–940 (2005).
Dombeck, D. A., Khabbaz, A. N., Collman, F., Adelman, T. L. & Tank, D. W. Imaging large-scale neural activity with cellular resolution in awake, mobile mice. Neuron. 56, 43–57 (2007).
Horton, N. G. et al. In vivo three-photon microscopy of subcortical structures within an intact mouse brain. Nat. Photon. 7, 205–209 (2013).
Sofroniew, N. J., Flickinger, D., King, J. & Svoboda, K. A large field of view two-photon mesoscope with subcellular resolution for in vivo imaging. eLife 5, e14472 (2016).
Ouzounov, D. G. et al. In vivo three-photon imaging of activity of GCaMP6-labeled neurons deep in intact mouse brain. Nat. Meth. 14, 388–390 (2017).
Papagiakoumou, E. et al. Scanless two-photon excitation of channelrhodopsin-2. Nat. Meth. 7, 848–854 (2010).
Packer, A. M., Russell, L. E., Dalgleish, H. W. P. & Häusser, M. Simultaneous all-optical manipulation and recording of neural circuit activity with cellular resolution in vivo. Nat. Meth. 12, 140–146 (2014).
Pnevmatikakis, E. A. et al. Simultaneous denoising, deconvolution, and demixing of calcium imaging data. Neuron 89, 285–299 (2016).
Yang, R., Weber, T. D., Witkowski, E. D., Davison, I. G. & Mertz, J. Neuronal imaging with ultrahigh dynamic range multiphoton microscopy. Sci. Rep. 7, 5817 (2017).
Desurvire, E., Giles, C. R. & Simpson, J. R. Gain saturation effects in high-speed, multichannel erbium-doped fiber amplifiers at lambda = 1.53 µm. J. Lightwave Tech. 7, 2095–2104 (1989).
Chen, T.-W. et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300 (2013).
Podgorski, K. & Ranganathan, G. Brain heating induced by near-infrared lasers during multiphoton microscopy. J. Neurophysiol. 116, 1012–1023 (2016).
Reddy, G. D., Kelleher, K., Fink, R. & Saggau, P. Three-dimensional random access multiphoton microscopy for functional imaging of neuronal activity. Nat. Neurosci. 11, 713–720 (2008).
Grewe, B. F., Langer, D., Kasper, H., Kampa, B. M. & Helmchen, F. High-speed in vivo calcium imaging reveals neuronal network activity with near-millisecond precision. Nat. Meth. 7, 399–405 (2010).
Katona, G. et al. Fast two-photon in vivo imaging with three-dimensional random-access scanning in large tissue volumes. Nat. Meth. 9, 201–208 (2012).
Chavarha, M. et al. Fast two-photon volumetric imaging of an improved voltage indicator reveals electrical activity in deeply located neurons in the awake brain. Preprint at bioRxiv https://doi.org/10.1101/445064 (2018).
Botcherby, E. J. et al. Aberration-free three-dimensional multiphoton imaging of neuronal activity at kHz rates. Proc. Natl Acad. Sci. USA 109, 2919–2924 (2012).
Weisenburger, S. et al. Volumetric Ca2+ imaging in the mouse brain using hybrid multiplexed sculpted light microscopy. Cell 177, 1050–1066 (2019).
Charan, K., Li, B., Wang, M., Lin, C. P. & Xu, C. Fiber-based tunable repetition rate source for deep tissue two-photon fluorescence microscopy. Biomed. Opt. Express 9, 2304–2311 (2018).
Acknowledgements
We thank members of the Xu research groups (T. Wang, D. Ouzounov, D. Sinfeld, C. Huang, X. Yang, A. Mok, F. Xia, Y. Hontani, Y. Qin, N. Akbari, K. Choe, T. Ciavatti and C. Zhuang) for their help. We thank M. Buttolph and L. G. Wright for discussions. This work is partially funded by NIH/NINDS (grant no. U01NS090530 to C.X.), and the National Science Foundation NeuroNex (grant no. DBI-1707312 to C.X.).
Author information
Authors and Affiliations
Contributions
C.X. initiated and supervised the study. B.L. designed and performed the experiments and analyzed the data. K.C. contributed to the construction of the fiber amplifiers. C.W. set up the awake imaging system and performed immunohistology and most of the animal surgeries. M.W. performed some of the animal surgeries. B.L. and C.X. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Rita Strack was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Demonstration of compensation of the gain transient in the EDFA.
a, A comparison of the amplified pulses before and after compensation. The envelope of the peaks of the pulses is shown. The pulse sequences correspond to one frame (512 × 512 pixels) of activity recording where only the somas are illuminated. b, Zoomed-in view of the input and output of the EDFA for a small segment of time (the blue region in a) after various numbers of iterations (indicated at the bottom of each panel) using the feedback loop. c, RMS fluctuation of the pulse intensity as a function of the number of iterations using the feedback loop. The insert is a zoomed-in view.
Supplementary Figure 2 The automatic feedback loop for compensation of the gain transient in the EDFA.
a, Diagram of the feedback loop. The electrical and optical paths are indicated in blue and red, respectively. b, The electrical (blue) and optical (red) waveforms at three positions indicated by (1), (2), and (3) in a. A detailed evolution of the waveforms is shown in Supplementary Video 1.
Supplementary Figure 3 Quantitative measurement of the neuron movement due to mouse motion.
a, Four neurons in a recording session of an awake mouse (Fig. 2) are shown and Supplementary Video 2. Scale bar, 20 μm. b, The measured movement of the center of the neuron in each frame and the average movement for the whole recording session. The RMS value of the neuron movement is about 1.2 μm, and the maximum movement is about 5 μm. Therefore, to capture the entire neuron, the radius of the ROI should be ~5 μm (or ~4 pixels for a FOV of 620 × 620 μm at 512 × 512 pixels) larger than the neuron.
Supplementary Figure 4 Spontaneous activity traces of all the neurons shown in Fig. 2.
a,b, Neurons are indexed in a, and the corresponding ROIs are shown in b. Scale bar, 100 μm. c, Spontaneous activity traces recorded from the indexed neurons (1–49), at a frame rate of 30 Hz. To the left of each trace is the index of the neuron. All traces were normalized to each individual baseline.
Supplementary Figure 5 60-minute recording of spontaneous activity of jRGECO1a-labeled neurons shown in Fig. 2 with 3PM.
Six consecutive recording sessions are shown with different colors. The duration of each recording session is 10 min (18,000 frames), limited by the file size of the acquisition software. The time gap between two successive recording sessions is less than 5 seconds.
Supplementary Figure 6 Longitudinal recording of neuronal activity at the same locations in an awake animal over 4 weeks.
Spontaneous activity traces from cortical neurons were recorded in an awake jRGECO1a mouse (12–17 weeks old, male) over a period of 4 weeks. High-resolution structural and ROI images of two recording sites at 615 μm and 575 μm are shown in the left and right panels. The activity traces of a fraction of neurons are shown in the middle panels, recorded at 30 Hz frame rate, and each was normalized to its baseline. The neurons are indexed alphabetically, and the same unique letter for each neuron is used in all imaging sessions. The days after the first imaging session, the age of the mouse, the average power used for activity recording and the imaging depth are indicated at the top of each image. The images have a FOV of 620 × 620 μm with 512 × 512 pixels per frame. Scale bars, 100 μm.
Supplementary Figure 7 Recording of spontaneous activity of jRGECO1a-labeled neurons at various depths of the same mouse after performing the chronic imaging shown in Supplementary Fig. 6.
High-resolution structural and ROI images of four recording sites at 575 μm, 615 μm, 900 μm and 1,000 μm are shown in the left panels. The activity traces of a fraction of neurons are shown in the right panels, recorded at 30-Hz frame rate, and each was normalized to its baseline. The neurons are indexed alphabetically. The days after the first imaging session, the age of the mouse, the average power used for activity recording and the imaging depth are indicated at the top of each image. The images have a FOV of 620 × 620 μm with 512 × 512 pixels per frame. Damage assessment of the imaged regions using immunohistology is shown in Supplementary Fig. 8b. Scale bars, 100 μm.
Supplementary Figure 9 Spontaneous activity traces of the neurons shown in Fig. 3.
a,b, Neurons are indexed in a, and the corresponding ROIs are shown in b. Scale bar, 100 μm. c, Spontaneous activity traces recorded from the indexed neurons (1–73) at a frame rate of 30 Hz. To the left of each trace is the index of the neuron. All traces were normalized to each individual baseline.
Supplementary Figure 10 In vivo 3PM of the red blood cell flow in the capillaries in a mouse brain (3 months old, similar data for n = 2 mice).
Texas red dextran was injected to label blood vessels. a, Three-dimensional reconstruction of images of blood vessels in the mouse cortex and the hippocampus (red, blood vessels; magenta, third-harmonic generation (THG) of external capsule (EC)). b, A selected frame in the EC. c, A structural image of capillaries located at 1,080 μm beneath the dura with a FOV of 220 × 220 μm, 512 × 512 pixels per frame, 30 frames per s, averaged over 100 s. d, Imaging of the capillaries (that is, the ROIs) using the AES with the same FOV and frame rate but no averaging. Red blood cells can be seen as shadows flowing through the fluorescent plasma. Two recording sessions are shown in Supplementary Videos 6 and 7. Scale bars, 50 μm.
Supplementary Figure 11 In vivo imaging of neurons labeled with YFP in a mouse brain, using the AES and a commercial MPM (FV1000MPE, Olympus).
a,b, Structural and ROI imaging of neurons (a) and dendrites (b). The blue dashed curves show the ROIs. The imaging depths are indicated at the top of the figures. Scale bars, 30 μm.
Supplementary information
Supplementary Information
Supplementary Figures 1–11 and Supplementary Notes 1 and 2
Supplementary Video 1
Compensation of the gain transient in the EDFA. a, Modulated optical pulses at the input of the EDFA. b, Amplified optical pulses at the output of the EDFA. c, RMS fluctuation of the pulse intensity as the function of the number of iterations of the feedback loop.
Supplementary Video 2
Measurement of the neuron movement due to mouse motion. The center of each neuron is marked with an X, and the ROIs are labelled with blue circles. The movement of the center in each frame and the average movement of the entire recording session is given in Supplementary Figure 3. The movie is sped up by 30 times, and the time is displayed on the top left corner. Scale bar, 20 µm.
Supplementary Video 3
ROI selection in the presence of animal motion; The ROIs are designed to be large enough so that the neurons remain within the ROIs even with the movement. For data 1 (2PM of the spontaneous activity of neurons) and 2 (3PM of the spontaneous activity of neurons), the ROIs occupy 3% and 5.5% of the FOV, respectively.
Supplementary Video 4
Three-photon imaging of the spontaneous activity of neurons labeled with jRGECO1a in an awake mouse. The movie was recorded with 512 × 512 pixels per frame and 30-Hz frame rate at the depth of 750 µm. The movie is sped up by 10 times, and the time is displayed on the top left corner. Scale bar, 100 µm.
Supplementary Video 5
Two-photon imaging of the spontaneous activity of neurons labeled with GCaMP6s in an awake mouse. The movie was recorded with 512 × 512 pixels per frame and 30-Hz frame rate at the depth of 680 µm. The movie is sped up by 20 times, and the time is displayed on the top left corner. Scale bar, 100 µm.
Supplementary Video 6
In vivo three-photon imaging of the red blood cell flow in the capillaries labeled with Texas red in CA1 region of a mouse brain. The movie was recorded with 512 × 512 pixels per frame and 30-Hz frame rate at the depth of 1,080 µm. The movie is not sped up, and the time is displayed on the top left corner. Scale bar, 50 µm.
Supplementary Video 7
In vivo three-photon imaging of the red-blood-cell flow in the capillaries labeled with Texas red in CA1 region of a mouse brain. The movie was recorded with 512 × 256 pixels per frame and 58-Hz frame rate at the depth of 1,080 µm. The movie is slowed down by two times, and the time is displayed on the top left corner. Scale bar, 50 µm.
Rights and permissions
About this article
Cite this article
Li, B., Wu, C., Wang, M. et al. An adaptive excitation source for high-speed multiphoton microscopy. Nat Methods 17, 163–166 (2020). https://doi.org/10.1038/s41592-019-0663-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-019-0663-9
This article is cited by
-
Live-cell imaging powered by computation
Nature Reviews Molecular Cell Biology (2024)
-
Large-scale cranial window for in vivo mouse brain imaging utilizing fluoropolymer nanosheet and light-curable resin
Communications Biology (2024)
-
Light-sheets and smart microscopy, an exciting future is dawning
Communications Biology (2023)
-
Real-time denoising enables high-sensitivity fluorescence time-lapse imaging beyond the shot-noise limit
Nature Biotechnology (2023)
-
Multiphoton intravital microscopy of rodents
Nature Reviews Methods Primers (2022)