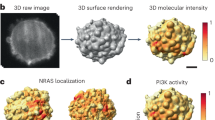
Abstract
Rapid developments in live-cell three-dimensional (3D) microscopy enable imaging of cell morphology and signaling with unprecedented detail. However, tools to systematically measure and visualize the intricate relationships between intracellular signaling, cytoskeletal organization and downstream cell morphological outputs do not exist. Here, we introduce u-shape3D, a computer graphics and machine-learning pipeline to probe molecular mechanisms underlying 3D cell morphogenesis and to test the intriguing possibility that morphogenesis itself affects intracellular signaling. We demonstrate a generic morphological motif detector that automatically finds lamellipodia, filopodia, blebs and other motifs. Combining motif detection with molecular localization, we measure the differential association of PIP2 and KrasV12 with blebs. Both signals associate with bleb edges, as expected for membrane-localized proteins, but only PIP2 is enhanced on blebs. This indicates that subcellular signaling processes are differentially modulated by local morphological motifs. Overall, our computational workflow enables the objective, 3D analysis of the coupling of cell shape and signaling.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Data are available from the corresponding author upon reasonable request.
Code availability
The latest version of the software described here, as well as a user’s guide, is available from https://github.com/DanuserLab/u-shape3D.
References
Munjal, A. & Lecuit, T. Actomyosin networks and tissue morphogenesis. Development 141, 1789–1793 (2014).
Rottner, K., Faix, J., Bogdan, S., Linder, S. & Kerkhoff, E. Actin assembly mechanisms at a glance. J. Cell Sci. 130, 3427–3435 (2017).
Simunovic, M., Voth, G. A., Callan-Jones, A. & Bassereau, P. When physics takes over: BAR proteins and membrane curvature. Trends Cell Biol. 25, 780–792 (2015).
Rangamani, P. et al. Decoding information in cell shape. Cell 154, 1356–1369 (2013).
Schmick, M. & Bastiaens, P. I. The interdependence of membrane shape and cellular signal processing. Cell 156, 1132–1138 (2014).
Planchon, T. A. et al. Rapid three-dimensional isotropic imaging of living cells using Bessel beam plane illumination. Nat. Methods 8, 417–U468 (2011).
Wu, Y. et al. Spatially isotropic four-dimensional imaging with dual-view plane illumination microscopy. Nat. Biotechnol. 31, 1032–1038 (2013).
Chen, B. C. et al. Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science 346, 1257998 (2014).
Dean, K. M., Roudot, P., Welf, E. S., Danuser, G. & Fiolka, R. Deconvolution-free subcellular imaging with axially swept light sheet microscopy. Biophys. J. 108, 2807–2815 (2015).
Welf, E. S. et al. Quantitative multiscale cell imaging in controlled 3D microenvironments. Dev. Cell 36, 462–475 (2016).
Fu, Q., Martin, B. L., Matus, D. Q. & Gao, L. Imaging multicellular specimens with real-time optimized tiling light-sheet selective plane illumination microscopy. Nat. Commun. 7, 11088 (2016).
Schink, K. O., Tan, K. W. & Stenmark, H. Phosphoinositides in control of membrane dynamics. Annu. Rev. Cell Dev. Biol. 32, 143–171 (2016).
Simanshu, D. K., Nissley, D. V. & McCormick, F. RAS proteins and their regulators in human disease. Cell 170, 17–33 (2017).
Driscoll, M. K. & Danuser, G. Quantifying modes of 3D cell migration. Trends Cell Biol. 25, 749–759 (2015).
Braunstein, M. L., Hoffman, D. D. & Saidpour, A. Parts of visual objects—an experimental test of the minima rule. Perception 18, 817–826 (1989).
Chazelle, B., Dobkin, D. P., Shouraboura, N. & Tal, A. Strategies for polyhedral surface decomposition: An experimental study. Comp. Geom. Theor. Appl. 7, 327–342 (1997).
Theisel, H., Rossi, C., Zayer, R. & Seidel, H. P. Normal based estimation of the curvature tensor for triangular meshes. In Proc. Pacific Conference on Computer Graphics and Applications 288–297 (IEEE, 2004).
Mangan, A. P. & Whitaker, R. T. Partitioning 3D surface meshes using watershed segmentation. IEEE Trans. Vis. Comp. Graph. 5, 308–321 (1999).
Kaick, O. V., Fish, N., Kleiman, Y., Asafi, S. & Cohen-Or, D. Shape segmentation by approximate convexity analysis. ACM Trans. Graph. 34, 1–11 (2014).
Singh, M., Seyranian, G. D. & Hoffman, D. D. Parsing silhouettes: the short-cut rule. Percept. Psychophys. 61, 636–660 (1999).
Dice, L. R. Measures of the amount of ecologic association between species. Ecology 26, 297–302 (1945).
Liu, T. L. et al. Observing the cell in its native state: imaging subcellular dynamics in multicellular organisms. Science 360, pii: eaaq1392 (2018).
Elliott, H. et al. Myosin II controls cellular branching morphogenesis and migration in three dimensions by minimizing cell-surface curvature. Nat. Cell Biol. 17, 137–147 (2015).
Zatulovskiy, E., Tyson, R., Bretschneider, T. & Kay, R. R. Bleb-driven chemotaxis of Dictyostelium cells. J. Cell Biol. 204, 1027–1044 (2014).
Cleveland, W. S. & Mcgill, R. Graphical perception and graphical methods for analyzing scientific data. Science 229, 828–833 (1985).
Sato, M. et al. Human lung epithelial cells progressed to malignancy through specific oncogenic manipulations. Mol. Cancer Res. 11, 638–650 (2013).
Redecke, V. et al. Hematopoietic progenitor cell lines with myeloid and lymphoid potential. Nat. Methods 10, 795–803 (2013).
Riedl, J. et al. Lifeact: a versatile marker to visualize F-actin. Nat. Methods 5, 605–607 (2008).
Leithner, A. et al. Diversified actin protrusions promote environmental exploration but are dispensable for locomotion of leukocytes. Nat. Cell Biol. 18, 1253–1259 (2016).
Johnson, H. W. & Schell, M. J. Neuronal IP3 3-kinase is an F-actin-bundling protein: role in dendritic targeting and regulation of spine morphology. Mol. Biol. Cell. 20, 5166–5180 (2009).
Yi, J., Wu, X. S., Crites, T. & Hammer, J. A. 3rd Actin retrograde flow and actomyosin II arc contraction drive receptor cluster dynamics at the immunological synapse in Jurkat T cells. Mol. Biol. Cell 23, 834–852 (2012).
Chu, J. et al. A bright cyan-excitable orange fluorescent protein facilitates dual-emission microscopy and enhances bioluminescence imaging in vivo. Nat. Biotechnol. 34, 760–767 (2016).
Stauffer, T. P., Ahn, S. & Meyer, T. Receptor-induced transient reduction in plasma membrane PtdIns(4,5)P2 concentration monitored in living cells. Curr. Biol. 8, 343–346 (1998).
Otsu, N. A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man, Cybern. 9, 62–66 (1979).
Desbrun, M., Meyer, M., Schroder, P. & Barr, A. H. Implicit fairing of irregular meshes using diffusion and curvature flow. In Proc. of the 26th Annual Conference on Computer Graphics and Interactive Techniques 317–324 (ACM Press/Addison-Wesley, 1999).
Aguet, F., Jacob, M. & Unser, M. Three-dimensional feature detection using optimal steerable filters. IEEE Image Proc. 2, 1158–1161 (2005).
Jacob, M. & Unser, M. Design of steerable filters for feature detection using Canny-like criteria. IEEE T. Pattern Anal. 26, 1007–1019 (2004).
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Goddard, T. D. et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
Kondor, R. I. & Lafferty, J. D. Diffusion kernels on graphs and other discrete input spaces. In Proc. International Conference on Machine Learning 315–322 (Morgan Kaufmann, 2002).
Kondor, R. & Vert, J. P. in Kernel Methods in Computational Biology (eds Schölkopf, B. et al) 171–192 (MIT Press, 2004).
Asafi, S., Goren, A. & Cohen-Or, D. Weak convex decomposition by lines-of-sight. Symp. Geom. 32, 23–31 (2013).
Möller, T. & Trumbore, B. Fast, minimum storage ray-triangle intersection. J. Graph. Tools 2, 21–28 (1997).
Christianini, N. & Shawe-Taylor, J. An Introduction to Support Vector Machines and Other Kernel-Based Learning Methods (Cambridge Univ. Press, 2000).
Breiman, L. Random forests. Mach. Learn. 45, 5–32 (2001).
Zhang, C. & Chen, T. Efficient feature extraction for 2D/3D objects in mesh representation. In Proc. 2001 International Conference on Image Processing 935–938 (IEEE, 2001).
Shapira, L., Shamir, A. & Cohen-Or, D. Consistent mesh partitioning and skeletonisation using the shape diameter function. Vis. Comput. 24, 249–259 (2008).
Suvrit, S. A short note on parameter approximation for von Mises–Fisher distributions: and a fast implementation of I s (x). Comput. Stat. 27, 177–190 (2012).
Machacek, M. & Danuser, G. Morphodynamic profiling of protrusion phenotypes. Biophys. J. 90, 1439–1452 (2006).
Harrower, M. & Brewer, C. A. ColorBrewer.org: an online tool for selecting colour schemes for maps. Cartogr. J. 40, 27–37 (2003).
Acknowledgements
This research was funded by grants from the Cancer Prevention Research Institute of Texas (nos. RR160057 to R.F. and R1225 to G.D.) and the National Institutes of Health (nos. F32GM116370 and K99GM123221 to M.K.D., K25CA204526 to E.S.W., F32GM117793 to K.M.D., R33CA235254 to R.F. and R01GM067230 to G.D.). Confocal imaging was performed at the UT Southwestern Live Cell Imaging Facility. Most surface renderings were performed using UCSF ChimeraX, which was developed by the Resource for Biocomputing, Visualization and Informatics at the University of California, San Francisco (supported by grant no. P41GM103311). We thank T. Goddard for assistance with ChimeraX, as well as I. de Vries, J. Renkawitz and M. Sixt for assistance differentiating dendritic cells. We would also like to thank F. Peri (University of Zurich) and members of her laboratory, especially M. Albert, for the unpublished images of microglia cells. We also thank P. Friedl (MD Anderson Cancer Center) for the MV3 melanoma cells, S. Morrison (UT Southwestern) for the primary melanoma cells, M. Sixt (IST Austria) for the dendritic cell precursors, J. Minna and J. Shay (UT Southwestern) for the transformed HBEC cells and R. McIntosh (University of Colorado, Boulder) for the U2OS osteosarcoma cells.
Author information
Authors and Affiliations
Contributions
M.K.D. and G.D. conceived and designed the study. M.K.D. wrote most of the software. A.R.J. developed the software’s graphical user interface. E.S.W. and M.K.D. performed most of the imaging experiments. K.M.D. and T.I. performed the U2OS experiments. K.M.D. and R.F. provided imaging resources. M.K.D. and G.D. wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information: Rita Strack was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 More views of the cells shown in Fig. 1a–f.
(a) Surface renderings of an HBEC expressing tractin-GFP. (b) MIPs of the cell shown in a. (c) Surface renderings of a dendritic cell expressing Lifeact-GFP. (d) Surface renderings of an MV3 melanoma cell expressing tractin-GFP. (e) MIPs of the cell shown in c. (f) MIPs of the cell shown in d.
Supplementary Figure 2 Variations in parameters governing deconvolution.
For the cell shown in Fig. 2f–p, a MIP (top) and the corresponding rendered surface colored by bleb detection (bottom) are shown. The blebs were detected using an SVM model derived from Weiner deconvolved images with an apodization height of 0.04. Images are (a) not deconvolved, Richardson-Lucy deconvolved for (b) 10 iterations and (c) 100 iterations, and Weiner deconvolved with an apodization height of (d) 0.00, (e) 0.04, (f) 0.08, (g) and 0.12.
Supplementary Figure 3 Segmentation of dendritic cells.
(a) A MIP across 4 µm of a deconvolved image of a dendritic cell expressing LifeAct-GFP. This image was segmented by processing the deconvolved image into three images that accentuate different features: (b) a renormalized deconvolved image, (c) an image that emphasizes the interior of the cell, and (d) an image that emphasizes planar features, such as lamellipodia. These three images were combined into (e) a composite image. (f) Finally, a mesh was generated from an isosurface of the composite image.
Supplementary Figure 4 Variations in parameters governing patch merging.
(a) Spill-depth based merging without triangle or line-of-sight (LOS) merging. A spill-depth ratio of 0.15 is used throughout the paper. (b) Triangle merging following spill-depth merging but without LOS merging. Elsewhere in the paper a triangle parameter of 0.7 is used. (c) LOS merging following spill-depth merging but without triangle merging. Elsewhere in the paper an LOS parameter of 0.7 is used.
Supplementary Figure 5 Additional example morphological motif detections.
Blebs are detected on two melanoma cells (left), filopodia on two HBEC cells (center) and lamellipodia on two dendritic cells (right).
Supplementary Figure 6 Validation of diverse bleb models.
(a) A melanoma cell with blebs detected via a model trained by a single user who clicked on patches that were certainly blebs or certainly not blebs. (b) Validation metrics for models trained by the same user on three different cells and applied to the cell shown in a. For the ‘Click on blebs’ column, training data was generated by asking the user to click on all patches that are blebs. For the ‘Click on not blebs’ column, the user was asked to click on all patches that are not blebs. Finally, for the ‘Click on certain’ column, the user was asked to click on all patches that are certainly blebs and then asked to click on all patches that are certainly not blebs. For this column, the user clicked on 39 of the 279 patches. (c) Models trained by the same user on four distinct MV3 cells (left), on those four cells combined (center), and on 19 MV3 cells (right). (d) Models trained by three different users on the same four MV3 cells as in c.
Supplementary Figure 7 Unsupervised clustering of patches and motifs.
(a) Supervised, SVM-based classification of patches into blebs (purple) and not blebs (gray). The SVM was trained on seven MV3 cells imaged via ASLM. (b) For the same set of cells, an unsupervised hierarchical clustering of patches into 2 clusters. (c) Unsupervised hierarchical clustering into 4 clusters of the supervised classification shown in a.
Supplementary Figure 8 Additional example morphological motif detections of cells imaged via diverse microscopy modalities.
(a) Three views of blebs detection on an MV3 cell expressing tractin-GFP imaged via a laser scanning confocal microscope. The bleb detection model was trained on eight MV3 cells imaged via the same microscope. (b) Extensions detected on microglia cells imaged within a zebrafish via a Zeiss LightSheet Z.1 light-sheet microscope. The extension detection model was trained on eight such cells. (c) Lamellipodia detected on a T cell imaged via lattice light-sheet microscopy. The lamellipodia detection model was trained on thirteen dendritic cells imaged via meSPIM.
Supplementary Figure 9 Workflow validation on confocal and ASLM acquired images.
MV3 cells expressing cytosolic GFP imaged via ASLM, a high-resolution light-sheet imaging modality, with blebs detected by a model trained on (a) nineteen MV3 cells expressing tractin-GFP imaged via meSPIM and (b) eight cells expressing cytosolic GFP imaged via ASLM. MV3 cells expressing tractin-GFP imaged via a laser scanning confocal microscope with blebs detected by a model trained on (c) nineteen MV3 cells expressing tractin-GFP imaged via meSPIM and (d) eight cells expressing tractin-GFP imaged via a confocal microscope. (e) Validation of bleb detection models trained on MV3 melanoma cells imaged via diverse microscopes (model column) and applied to the same or different sets of MV3 melanoma cell images (data column).
Supplementary Figure 10 Uropod and retraction fiber detection.
(a) Blebs detected on the MV3 cell shown in Fig. 6b, which is expressing GFP-KrasV12, using a model trained on MV3 cells expressing tractin-GFP. Each bleb is randomly colored. (b) Uropods and retraction fibers detected on MV3 cells expressing GFP-KrasV12. (c) The uropod detected on the cell shown in a. (d) For the same cell, detected blebs (see a) with the detected uropod patches (see c) removed.
Supplementary Figure 11 Measuring intensity surface localization.
(a) A surface rendering of an MV3 melanoma cell with detected blebs shown white and detected non-blebs shown black. (b) A surface rendering of the same cell colored by normalized local bleb density. White indicates regions of high local bleb density, whereas black indicates regions of low local bleb density. Distributions of normalized bleb density for cells expressing (c) GFP-KrasV12 (n=13 cells) and (d) PLCΔ-PH-GFP (n=6 cells), a PIP2 translocation biosensor. The red lines show the distribution above one standard deviation above the mean intensity, and the orange lines shows the distribution below one standard deviation below the mean intensity. (e) Fluorescence intensity at mesh faces vs. distance from a bleb edge for synthetic images. The dark blue line shows the intensity for 9 cytosolically labeled cells, and the lighter blue lines show the intensity for the same set of 9 cells with cortical, rather than cytosolic, labeling of various thicknesses. (f) Distributions of fluorescence intensity, measured over 1 µm, for 5 GFP cytosolically labeled MV3 cells. The solid line shows the intensity distribution for mesh faces on blebs, whereas the dashed line shows the distribution off blebs. The blue line shows the distribution for mesh faces on blebs with greater than mean bleb volume, whereas the red line shows the distribution on blebs with less than mean volume.
Supplementary Figure 12 Characterization of Wave2 and cofilin-1 knockout cells.
Total cell lysates (20 µg) of U2OS wild-type, Wave2 and cofilin-1 knockout cells were separated by SDS-PAGE and immunoblotted as indicated. GAPDH served as loading control. This blot is representative of two blots (n=2).
Supplementary Figure 13 Segmentation assessment.
An MV3 melanoma cell expressing tractin-GFP. (a) Five xy-slices sampled 4 µm apart. The raw data is shown cyan, whereas a pixelation of the mesh representing the cell surface is shown red. (b) A 3D rendering of the extracted cell surface for the same cell.
Supplementary Figure 14 Mutual visibility and motion.
(a) The convergence of the mutual visibility between patches as a function of number of rays tested. The black line shows the standard error about the mean mutual visibility, and the gray region shows the standard deviation of that standard error. Throughout the paper 20 rays are used to calculate mutual visibility. (b) The frequency of protrusive motion minus the frequency of retractive motion at mesh faces on and off blebs as a function of surface speed. The ‘backwards’ motion algorithm, used elsewhere in the paper, is shown on the left, and the ‘forwards’ motion algorithm is shown on the right.
Supplementary Figure 15 Analysis of synthetic images of blebby cells.
(a) Segmentation of the cell surface into convex patches via the same workflow used for microscopic data. (b) Detection of blebs using a single SVM bleb classifier trained on synthetic data.
Supplementary information
Supplementary Information
Supplementary Figs. 1–15 and Supplementary Tables 1–6.
Supplementary Video 1
Rotations of surface renderings. Renderings of the extracted surfaces of a dendritic cell (first), an MV3 melanoma cell (second) and a transformed human bronchial epithelial cell (third).
Supplementary Video 2
3D image of an MV3 melanoma cell expressing GFP-KrasV12. The image’s field of view is 79 × 47 × 55 µm3 in x, y and z, respectively, and the video scans through the z-dimension.
Supplementary Video 3
Video of an xy-slice of an MV3 melanoma cell expressing GFP-KrasV12. The field of view is 27 × 32 µm2, and the time between frames is 21 s.
Supplementary Video 4
Rotations of morphological motif detections.Blebs detected on an MV3 melanoma cell (first), lamellipodia detected on a dendritic cell (second) and filopodia detected on a human bronchial epithelial cell (third).
Supplementary Video 5
Extensions detected on an MDA-MB-231 human breast cancer cell moving through the vasculature of a zebrafish embryo. The cell was imaged via an adaptive optics lattice light-sheet microscope.
Rights and permissions
About this article
Cite this article
Driscoll, M.K., Welf, E.S., Jamieson, A.R. et al. Robust and automated detection of subcellular morphological motifs in 3D microscopy images. Nat Methods 16, 1037–1044 (2019). https://doi.org/10.1038/s41592-019-0539-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-019-0539-z
This article is cited by
-
Bleb protrusions help cancer cells to cheat death
Nature (2023)
-
Cellular harmonics for the morphology-invariant analysis of molecular organization at the cell surface
Nature Computational Science (2023)
-
A deep learning framework for quantitative analysis of actin microridges
npj Systems Biology and Applications (2023)
-
Bridging live-cell imaging and next-generation cancer treatment
Nature Reviews Cancer (2023)
-
Blebs promote cell survival by assembling oncogenic signalling hubs
Nature (2023)