Abstract
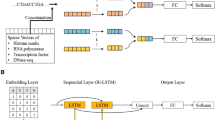
The decoding of transcription factor (TF) binding signals in genomic DNA is a fundamental problem. Here we present a prediction model called BindSpace that learns to embed DNA sequences and TF labels into the same space. By training on binding data from hundreds of TFs and embedding over 1 M DNA sequences, BindSpace achieves state-of-the-art multiclass binding prediction performance, in vitro and in vivo, and can distinguish between signals of closely related TFs.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
We used only public data sets in this study. Each data set can be accessed in the original publication referenced.
Code availability
Open source code for BindSpace as well as the trained BindSpace model are freely available for download (https://bitbucket.org/hy395/selex_embed for source code for training BindSpace; https://bitbucket.org/hy395/bindspace for R package to make predictions with the trained model).
References
Berger, M. F. et al. Nat. Biotechnol. 24, 1429–1435 (2006).
Warren, C. L. et al. Proc. Natl Acad. Sci. 103, 867–872 (2006).
Gordân, R. et al. Cell Rep. 3, 1093–1104 (2013).
Maerkl, S. J. & Quake, S. R. Science 315, 233–237 (2007).
Jolma, A. et al. Cell 152, 327–339 (2013).
Ghandi, M., Lee, D., Mohammad-Noori, M. & Beer, M. A. PLoS Comput. Biol. 10, e1003711 (2014).
Setty, M. & Leslie, C. S. PLoS Comput. Biol. 11, e1004271 (2015).
Yang, L. et al. Mol. Syst. Biol. 13, 1–14 (2017).
Shen, N. et al. Cell Syst. 6, 470–483.e8 (2018).
Wu, L. et al. StarSpace: Embed All The Things! In 32nd AAAI Conference on Artificial Intelligence (2018).
Melvin, I., Weston, J., Noble, W. S. & Leslie, C. PLoS Comput. Biol. 7, e1001047 (2011).
Dai, H. et al. Bioinformatics 15, 3575–3583 (2017).
Grant, C. E., Bailey, T. L. & Noble, W. S. Bioinformatics 27, 1017–1018 (2011).
Alipanahi, B., Delong, A., Weirauch, M. T. & Frey, B. J. Nat. Biotechnol. 33, 831–838 (2015).
Barrera, L. A. et al. Science 351, 1450–1454 (2016).
Wei, G. H. et al. EMBO J. 29, 2147–2160 (2010).
Berger, M. F. et al. Cell 133, 1266–1276 (2008).
Slattery, M. et al. Cell 147, 1270–1282 (2011).
Morgulis, A., Gertz, E. M., Schäffer, A. A. & Agarwala, R. J. Comput. Biol. 13, 1028–1040 (2006).
Simon, N., Friedman, J., Hastie, T. & Tibshirani, R. J. Stat. Softw. 39, 1–13 (2011).
van Der Maaten, L. & Hinton, G. J. Mach. Learn. Res. 9, 2579–2605 (2008).
van der Maaten, L. J. Mach. Learn. Res. 15, 3221–3245 (2014).
Krijthe, J. H. Rtsne: T-distributed stochastic neighbor embedding using Barnes-Hut implementation. https://github.com/jkrijthe/Rtsne (2015).
Litzenburger, U. M. et al. Genome Biol. 18, 15 (2017).
Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y. & Greenleaf, W. J. Nat. Methods 10, 1213–1218 (2013).
Mariani, L., Weinand, K., Vedenko, A., Barrera, L. A. & Bulyk, M. L. Cell Syst. 5, 187–201.e7 (2017).
Acknowledgements
We thank J. Weston for suggesting the StarSpace algorithm and providing access to his code. We also thank W. Stafford Noble, J. Bilmes, and J. Schreiber for helpful comments on the project. This work was supported by NIH/NHGRI U01 award HG009395. H.Y. was also supported by the Tri-Institutional Training Program in Computational Biology and Medicine.
Author information
Authors and Affiliations
Contributions
H.Y. developed and implemented the model, performed all computational experiments and co-wrote the manuscript. M.K. helped to perform quality control, initial benchmarking, visualization and formulation of evaluations. L.Z. advised on quality control and experimented with alternative embedding ideas. Y.L. helped with visualization and advised on analyses. C.S.L. supervised the research, contributed to the model design and co-wrote the manuscript. All authors discussed the results and contributed to the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information: Nicole Rusk was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 t-SNE visualization of the embedding space.
a) Colored points represent embedding of enriched probes from HT-SELEX experiments (n=50 from each experiment). Black points and labels represent embedding of 171 TFs (except for Homeodomain TFs). 98 Homeodomain TFs are shown by gray points, and the associated labels are not shown because there are too many of them. Red points and labels represent embedding of 13 major families (families with more than three members). b) Zooming into top left corner of a) to look at the bZIP family. Colored points represent embedding of enriched probes from bZIP HT-SELEX experiments. Black points and labels represent embedding of TFs. Red point and label represent embedding of bZIP family label.
Supplementary Figure 2 Sequence logos for selected BindSpace TFs.
We generated a sequence logo for each TF in BindSpace through alignment of 10-mers embedded near to each model vector (Methods). Sequence logos with at least one informative nucleotide (information content greater than 1) are shown here.
Supplementary Figure 3 Multiclass prediction performance of six models on held-out HT-SELEX probes.
BindSpace, BindSpace+ and LASSO (one-versus-all) are trained as multiclass classifiers; DeepBind, LASSO_binary, and PWM are trained as binary classifiers (Methods). Classification performance is measured by F1 score (left) and auPR (right) for each of 243 TF classes. Boxplots show median, upper and lower quartiles, and highest and lowest values excluding outliers.
Supplementary Figure 4 Confusion matrices for BindSpace, PWM, LASSO and DeepBind on held-out HT-SELEX probes.
Multiclass prediction performance for BindSpace (first column), PWM (second column), LASSO (third column) and DeepBind (fourth column) as shown by confusion matrices normalized by class size for all 13 major families. Rows are true labels, and columns are predicted labels. See Supplementary Note for confusion matrices for other families.
Supplementary Figure 5 Performance evaluation of BindSpace, PWM, LASSO and DeepBind on BAR15A PBM data.
For each of the 14 homeodomain PBM experiments that are represented in both BAR15A and BindSpace, we predicted the binding affinity of TFs to the top n=3000 probes with highest intensity from each PBM experiment using BindSpace (first row), PWM (second row), LASSO (third row) and DeepBind (fourth row). In each plot, we show the Spearman’s correlation of estimated affinities to measured PBM intensities. For 8 of the 14 TFs, the PBM intensity correlated best with the BindSpace model for the same TF (marked with *), compared to 7/14 TFs for LASSO and PWMs and 6/14 TFs for DeepBind.
Supplementary Figure 6 Performance comparison of BindSpace, PWM, LASSO and DeepBind for binary ENCODE ChIP-seq peaks versus background sequence task.
Performance of BindSpace, PWM, LASSO and DeepBind on the task of distinguishing n=39 TF ChIP-seq peaks from flanks (top) and ChIP-seq peaks from dinucleotide shuffled peak sequences (bottom), evaluated by F1 score (left) and auROC (right). One-sided Wilcoxon signed rank test is used when the direction is clear, otherwise two-sided test is used (noted in the figure). Boxplots show median, upper and lower quartiles, and highest and lowest values excluding outliers.
Supplementary Figure 7 Performance comparison of BindSpace, PWM, LASSO and DeepBind for binary task of distinguishing TF-occupied ATAC-seq peaks versus TF-unoccupied ATAC-seq peaks.
Performance of BindSpace, PWM, LASSO and DeepBind on the task of distinguishing TF-occupied ATAC-seq peaks versus TF-unoccupied ATAC-seq peaks in K562 (top, n=17 TFs) and GM12878 (bottom, n=11 TFs) as evaluated by F1 score (left) and auROC (right). One-sided Wilcoxon signed rank test is used when the direction is clear, otherwise two-sided test is used (noted in the figure). Boxplots show median, upper and lower quartiles, and highest and lowest values excluding outliers.
Supplementary Figure 8 Performance of PWM, LASSO and DeepBind for distinguishing CEBPB specific peaks versus CEBPG specific peaks in HepG2.
From top to bottom, we show the results for PWM, LASSO and DeepBind. From left to right for each method, we show the predicted scores of CEBPB specific peaks (n=3494), CEBPG specific peaks (n=3494), and common peaks (n=6506) in the scatter plots. One-sided Wilcoxon signed-rank test P values are reported in the top right corner. Each probe is assigned to be CEBPB specific (red), CEBPG specific (blue) or low affinity (gray). See Methods for empirical thresholds for predicting low affinity (negative) probes for each method. Pie charts show the proportion of CEBPB specific peaks versus CEBPG specific peaks in each group.
Supplementary Figure 9 Performance of BindSpace, PWM, LASSO and DeepBind for distinguishing NR2F6 specific peaks versus NR2F1 specific peaks in HepG2.
From top to bottom, we show the results for BindSpace, PWM, LASSO and DeepBind. From left to right for each method, we show the predicted scores of NR2F6 specific peaks (n=3819), NR2F1 specific peaks (n=3819), and common peaks (n=6188) in the scatter plots. One-sided Wilcoxon signed-rank test P values are reported in the top right corner. Each probe is assigned to be NR2F6 specific (red), NR2F1 specific (blue) or low affinity (gray). See Methods for empirical thresholds for predicting low affinity (negative) probes for each method. Pie charts show the proportion of NR2F6 specific peaks versus NR2F1 specific peaks in each group.
Supplementary Figure 10 Multiclass prediction performance of TF binding at bZIP ChIP-seq peaks in HepG2 cell line.
Multiclass classification performance of BindSpace, PWM, LASSO and DeepBind for bZIP family members in the HepG2 cell line. For each plot, we performed multiclass classification using BindSpace on the top 10,000 peaks for bZIP TF ChIP-seq and show the proportion of predicted labels for each model. BindSpace successfully ranked the ChIP-ed TF above other family members in 5 out of 7 cases, as indicated by *, as compared to 3 out of 7 for PWM, 3 out of 7 for LASSO, 2 out of 7 for DeepBind.
Supplementary Figure 11 Multiclass prediction performance of TF binding for the bZIP ChIP-seq combined atlas.
Performance comparison for multiclass classification of bZIP family TF ChIP-seq peaks evaluated by one-versus-all auPR scores (top left), one-versus-all F1 scores (top right) and confusion matices (bottom).
Supplementary Figure 12 BindSpace allows multi-label prediction of ATAC-seq peaks with high resolution.
ChIP-seq signals (ENCODE signal p-value track) and ATAC-seq signals (RPM) (top), BindSpace predictions (middle) and BindSpace+ predictions (bottom) at two different ATAC-seq peaks in K562.
Supplementary information
Supplementary Information
Supplementary Figures 1–12 and Supplementary Note.
Supplementary Table 1
List of top 200 8-mers that are closest to each TF label in BindSpace.
Supplementary Table 2
Performance overview of BindSpace, PWM, one-versus-all LASSO and DeepBind on all evaluation tasks.
Supplementary Table 3
Information on 270 HT-SELEX experiments used for training and number of unique and non-unique probes among top 2,000 intensity probes in each experiment.
Source data
Rights and permissions
About this article
Cite this article
Yuan, H., Kshirsagar, M., Zamparo, L. et al. BindSpace decodes transcription factor binding signals by large-scale sequence embedding. Nat Methods 16, 858–861 (2019). https://doi.org/10.1038/s41592-019-0511-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-019-0511-y
This article is cited by
-
SIMBA: single-cell embedding along with features
Nature Methods (2023)
-
BindVAE: Dirichlet variational autoencoders for de novo motif discovery from accessible chromatin
Genome Biology (2022)
-
Prediction of protein–ligand binding affinity from sequencing data with interpretable machine learning
Nature Biotechnology (2022)