Abstract
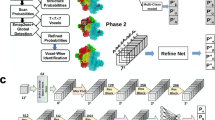
Although structures determined at near-atomic resolution are now routinely reported by cryo-electron microscopy (cryo-EM), many density maps are determined at an intermediate resolution, and extracting structure information from these maps is still a challenge. We report a computational method, Emap2sec, that identifies the secondary structures of proteins (α-helices, β-sheets and other structures) in EM maps at resolutions of between 5 and 10 Å. Emap2sec uses a three-dimensional deep convolutional neural network to assign secondary structure to each grid point in an EM map. We tested Emap2sec on EM maps simulated from 34 structures at resolutions of 6.0 and 10.0 Å, as well as on 43 maps determined experimentally at resolutions of between 5.0 and 9.5 Å. Emap2sec was able to clearly identify the secondary structures in many maps tested, and showed substantially better performance than existing methods.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The raw data of accuracies are provided in Supplementary Tables 1, 3 and 4. The experimental EM maps can be downloaded from EMDB (Supplementary Table 3). Output files from Emap2sec for the simulated and experimental maps that support the findings of this study are available from the corresponding author upon request.
Code availability
The Emap2sec program is freely available for academic use through Code Ocean33 and via http://www.kiharalab.org/emap2sec/index.html and https://www.github.com/kiharalab/Emap2sec. Simulated maps are available in the Code Ocean code capsule.
References
Kuhlbrandt, W. Cryo-EM enters a new era. eLife 3, e03678 (2014).
Cheng, Y. Single-particle cryo-EM—how did it get here and where will it go. Science 361, 876–880 (2018).
Patwardhan, A. Trends in the Electron Microscopy Data Bank (EMDB). Acta Crystallogr. D Struct. Biol. 73, 503–508 (2017).
Nogales, E. The development of cryo-EM into a mainstream structural biology technique. Nat. Methods 13, 24–27 (2016).
Esquivel-Rodriguez, J. & Kihara, D. Computational methods for constructing protein structure models from 3D electron microscopy maps. J. Struct. Biol. 184, 93–102 (2013).
Kirmizialtin, S., Loerke, J., Behrmann, E., Spahn, C. M. & Sanbonmatsu, K. Y. Using molecular simulation to model high-resolution cryo-EM reconstructions. Methods Enzym. 558, 497–514 (2015).
Miyashita, O., Kobayashi, C., Mori, T., Sugita, Y. & Tama, F. Flexible fitting to cryo-EM density map using ensemble molecular dynamics simulations. J. Comput. Chem. 38, 1447–1461 (2017).
Esquivel-Rodriguez, J. & Kihara, D. Fitting multimeric protein complexes into electron microscopy maps using 3D zernike descriptors. J. Phys. Chem. B 116, 6854–6861 (2012).
Saha, M. & Morais, M. C. FOLD-EM: automated fold recognition in medium- and low-resolution (4–15 Å) electron density maps. Bioinformatics 28, 3265–3273 (2012).
Zheng, W. Accurate flexible fitting of high-resolution protein structures into cryo-electron microscopy maps using coarse-grained pseudo-energy minimization. Biophys. J. 100, 478–488 (2011).
Brown, A. et al. Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr. D Biol. Crystallogr. 71, 136–153 (2015).
Terwilliger, T. C. et al. Iterative model building, structure refinement and density modification with the PHENIX auto build wizard. Acta Crystallogr. D Biol. Crystallogr. 64, 61–69 (2008).
DiMaio, F. et al. Atomic-accuracy models from 4.5-Å cryo-electron microscopy data with density-guided iterative local refinement. Nat. Methods 12, 361–365 (2015).
Chen, M., Baldwin, P. R., Ludtke, S. J. & Baker, M. L. De novo modeling in cryo-EM density maps with pathwalking. J. Struct. Biol. 196, 289–298 (2016).
Terashi, G. & Kihara, D. De novo main-chain modeling for EM maps using MAINMAST. Nat. Commun. 9, 1618 (2018).
Terashi, G. & Kihara, D. De novo main-chain modeling with MAINMAST in 2015/2016 EM model challenge. J. Struct. Biol. 204, 351–359 (2018).
Jiang, W., Baker, M. L., Ludtke, S. J. & Chiu, W. Bridging the information gap: computational tools for intermediate resolution structure interpretation. J. Mol. Biol. 308, 1033–1044 (2001).
Dou, H., Burrows, D. W., Baker, M. L. & Ju, T. Flexible fitting of atomic models into cryo-EM density maps guided by helix correspondences. Biophys. J. 112, 2479–2493 (2017).
Kong, Y., Zhang, X., Baker, T. S. & Ma, J. A structural-informatics approach for tracing β-sheets: building pseudo-Cα traces for β-strands in intermediate-resolution density maps. J. Mol. Biol. 339, 117–130 (2004).
Si, D. & He, J. Modeling β-traces for β-barrels from cryo-EM density maps. Biomed. Res. Int. 2017, 1793213 (2017).
Si, D. & He, J. Tracing β strands using StrandTwister from cryo-EM density maps at medium resolutions. Structure 22, 1665–1676 (2014).
Lindert, S. et al. EM-fold: de novo atomic-detail protein structure determination from medium-resolution density maps. Structure 20, 464–478 (2012).
Berman, H. M. et al. The Protein Data Bank. Nucleic Acids Res. 28, 235–242 (2000).
Biswas, A. et al. An effective computational method incorporating multiple secondary structure predictions in topology determination for cryo-EM images. IEEE/ACM Trans. Comput. Biol. Bioinform 14, 578–586 (2017).
Li, R. J., Si, D., Zeng, T., Ji, S. W. & He, J. Deep convolutional neural networks for detecting secondary structures in protein density maps from cryo-electron microscopy. in 2016 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) (eds Tian, T. et al.) 41–46 (IEEE, 2016).
Russakovsky, O. et al. Image net large scale visual recognition challenge. Int. J. Comput. Vis. 115, 1–42 (2015).
Maturana, D. & Scherer, S. VoxNet: a 3D convolutional neural network for real-time object recognition. in 2015 IEEE/RSJ International Conference on Intelligent Robots and Systems (IROS) 922–928 (IEEE, 2015).
Jones, D. T. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292, 195 (1999).
Fox, N. K., Brenner, S. E. & Chandonia, J. M. SCOPe: structural classification of proteins—extended, integrating SCOP and ASTRAL data and classification of new structures. Nucleic Acids Res. 42, D304–D309 (2014).
Deneka, D., Sawicka, M., Lam, A. K. M., Paulino, C. & Dutzler, R. Structure of a volume-regulated anion channel of the LRRC8 family. Nature 558, 254–259 (2018).
Langer, G., Cohen, S. X., Lamzin, V. S. & Perrakis, A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 3, 1171–1179 (2008).
Tang, G. et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007).
Subramaniya, S. R. M. V., Terashi, G. & Kihara, D. Protein secondary structure detection in intermediate resolution cryo-electron microscopy maps using deep learning v.2.0 (Code Ocean, 2019); https://doi.org/10.24433/CO.3068754.v2
Frishman, D. & Argos, P. Knowledge-based protein secondary structure assignment. Proteins 23, 566–579 (1995).
Monroe, L., Terashi, G. & Kihara, D. Variability of protein structure models from electron microscopy. Structure 25, 592–602 (2017).
Acknowledgements
The authors acknowledge C. Christoffer for his help in finalizing the manuscript. This work was partly supported by the National Institutes of Health (R01GM123055), the National Science Foundation (DMS1614777 and CMMI1825941) and the Purdue Institute of Drug Discovery.
Author information
Authors and Affiliations
Contributions
D.K. conceived the study. S.R.M.V.S. designed the Emap2sec architecture with D.K. and G.T., and S.R.M.V.S. implemented it. The datasets were selected by S.R.M.V.S. and G.T. The experiments were designed by S.R.M.V.S. and D.K., and were carried out by S.R.M.V.S. S.R.M.V.S., G.T. and D.K. analyzed the results. The manuscript was drafted by S.R.M.V.S. D.K. administrated the project and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information: Allison Doerr was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Performance comparison with networks trained on a larger dataset.
The performance of secondary structure detection reported in the main text was computed with networks trained on 63 maps as described in the Method section. Here we compared with the performance using networks trained on a larger training dataset, 982 maps for the phase 1 and 981 maps for the phase 2 network (y-axis). These are all the remaining maps from the 2000 non-redundant maps minus the 34 testing maps and 3 maps which caused errors when voxel data were generated. See Supplementary Table 2 for average accuracy values of the measures shown in the plots.
Supplementary Figure 2 Structure detection accuracy for experimental EM maps using both experimental maps and simulated maps as training data.
For each experimental EM map, the structure detection result using the network trained on the experimental maps (x-axis) and the result obtained when trained on both experimental maps and simulated maps (y-axis) was plotted. Left, overall voxel-based F1-score; right, overall voxel-based accuracy. The average F1-score: 0.415 (trained on experimental maps); 0.402 (trained on both experimental and simulated maps). The average accuracy: 0.422 (trained on experimental maps); 0.421 (trained on both experimental and simulated maps)
Supplementary Figure 3 Examples of detected secondary structures that can help structure assignment.
EMD-8693 is a map of BG505 SOSIP.664 (HIV capsid proteins) in complex with antibodies, which was determined at 6.2 Å resolution. (This map is also shown in Fig. 5b). The complex (PDB ID: 5viy) has six different chains. Among six, two are HIV capsid proteins and the rest are Fab heavy and light chains. The detected β structures locating at the three edges of the map in the figure (one of them is marked with a red circle) would help users to assign Fab proteins, which have β-sandwich structures, to the map density. In addition, α helices are clearly visible at the center of the map in the figure, which apparently correspond to chain A, B, C, the only proteins of the α-class in this complex. EMD-3329 is a 6.1 Å map of the complex of Ser/Thr kinase Tor2 and target of rapamycin complex subunit Lst8 (PDB ID: 5fvm). There are 4 chains in this complex. Chain A and B are Tor2, 2471 residue-long α-class structures. Chain C and D are LST8, 303 residue-long β-class structures (marked with circles). The locations of the two round-shaped β-class structures of LST8 are clearly identifiable in the Emap2sec’s output (right panel). For this EM map, there is a misrecognized β strands region at the bottom of the figure, but the shape and the volume of the region does not fit well to the structure of LST8.
Supplementary Figure 4 Examples of structure detection by Phenix and ARP/wARP.
Left, structure detection in a simulated map at 6.0 Å of d1kafa. Yellow and pink, β strands and helices, respectively, of the crystal structure; orange, helices detected by Phenix; blue, helices detected by ARP/wARP. The simulated map density was omitted from the figure for clarity. Phenix correctly identified two helices, but missed the one on the top, and over predicted one helix for two β strands at the bottom. ARP/wARP recognized the helix on the top, and a short fragment of helix for the helix at the bottom. As shown in Supplementary Table 4, Emap2sec recognized all the secondary structures, the three helices and six β strands, correctly. Right, a simulated map at 6.0 Å of d1a12a_. Three panels are shown. The middle one is the crystal structure; The left one is structures detected by ARP/wARP. The right one shows structures detected by Phenix. As shown for this β-class protein, both methods did not show meaningful results. Phenix output many α helices, which were obviously incorrect. ARP/wARP output short fragments and did not detect any secondary structures. In contrast, Emap2sec correctly detected 26 β strands out of 29. For the 10.0 Å map of this protein, Emap2sec still detected 25 β strands out of 29 correctly. On the other hand, Phenix showed an error message, which says “unable to build secondary structure model”. ARP/wARP output small fragment structures as it did for the 6.0 Å map.
Supplementary information
Supplementary Information
Supplementary Figs. 1–4 and Supplementary Tables 2, 5 and 6
Supplementary Table 1
Detailed results of the secondary structure detection on the simulated EM maps. Results of all evaluation measures, the voxel-wise F1 score, accuracy, the residue-based Q3 score and the segment-based accuracy of each map at 6.0 Å and 10.0 Å are provided. N/A in the table indicates that the secondary structure does not exist in the structure according to STRIDE.
Supplementary Table 3
Detailed results of the secondary structure detection on the dataset of experimental EM maps. Notations are the same as in Supplementary Table 1.
Supplementary Table 4
Performance comparison with Phenix and ARP/wARP. The segment-based accuracies of α-helices and β-strands were compared on the dataset of simulated maps at 6.0 Å and 10.0 Å. Total, the total number of the secondary structure segments in the protein; N/A, no segments of the type existed in the protein; ERROR, the program showed an error message and did not output detected segments in the PDB file. A segment was considered as correctly detected if at least 50% of the voxels in that segment had the correctly assigned class label. The counts show the number of correctly identified segments (that is, over predictions were not counted).
Supplementary Table 7
The list of SCOPe IDs of the 2,000 simulated maps.
Rights and permissions
About this article
Cite this article
Maddhuri Venkata Subramaniya, S.R., Terashi, G. & Kihara, D. Protein secondary structure detection in intermediate-resolution cryo-EM maps using deep learning. Nat Methods 16, 911–917 (2019). https://doi.org/10.1038/s41592-019-0500-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-019-0500-1
This article is cited by
-
CryoREAD: de novo structure modeling for nucleic acids in cryo-EM maps using deep learning
Nature Methods (2023)
-
CR-I-TASSER: assemble protein structures from cryo-EM density maps using deep convolutional neural networks
Nature Methods (2022)
-
Residue-wise local quality estimation for protein models from cryo-EM maps
Nature Methods (2022)
-
Extraction of protein dynamics information from cryo-EM maps using deep learning
Nature Machine Intelligence (2021)
-
DeepEMhancer: a deep learning solution for cryo-EM volume post-processing
Communications Biology (2021)