Abstract
Chemical inhibitors have revealed requirements for protein synthesis that drive cellular plasticity. We developed a genetically encodable protein synthesis inhibitor (gePSI) to achieve cell-type-specific temporal control of protein synthesis. Controlled expression of the gePSI in neurons or glia resulted in rapid, potent and reversible cell-autonomous inhibition of protein synthesis. Moreover, gePSI expression in a single neuron blocked the structural plasticity induced by single-synapse stimulation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
Source data for Figs. 1–3 are available online. The full vector sequences used in this study have been submitted to the NIH genetic sequence database (GenBank). The GenBank accession numbers are listed below: MN224153 (pCAMKIIa-TetOn3G), MN224154 (pCMV-FLEx-TetOn3G-3xHA-FLEx), MN224155 (pGFAP-TetOn3G), MN224156 (pTRE3G-Bi-a-Chain), MN224157 (pTRE3G-Bi-b-Chain), MN224158 (pTRE3G-Bi-gePSI-pCamk2a-AcGFP), MN224159 (pTRE3G-Bi-gePSI-pCMV-AcGFP), MN224160 (pTRE3G-Bi-gePSI-pGFAP-AcGFP), MN224161 (pTRE3G-Bi-ProRIP). The following plasmids have been deposited on Addgene (Addgene ID): pTRE3G-Bi-gePSI-pCMV-AcGFP (131008), pCAMKIIa-TetOn3G (133727), pGFAP-TetOn3G (133728), pCMV-FLEx-TetOn3G-3xHA-FLEx (133729), pTRE3G-Bi-α-chain (133730), pTRE3G-Bi-β-chain (133731), pTRE3G-Bi-gePSI-pCamk2a-AcGFP (133732), pTRE3G-Bi-gePSI-pGFAP-AcGFP (133733). Further data can be requested from the corresponding author upon reasonable request.
Change history
25 October 2019
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
References
Sutton, M. A. & Schuman, E. M. Cell 127, 49–58 (2006).
Walsh, M. J., Dodd, J. E. & Hautbergue, G. M. Virulence 4, 774–784 (2013).
Puri, M., Kaur, I., Perugini, M. A. & Gupta, R. C. Drug Discov. Today 17, 774–783 (2012).
Endo, Y. & Tsurugi, K. Nucleic Acids Symp. Ser. 262, 139–142 (1988).
Hey, T. D., Hartley, M. & Walsh, T. A. Plant Physiol. 107, 1323–1332 (1995).
Mak, A. N.-S. et al. Nucleic Acids Res. 35, 6259–6267 (2007).
Schmidt, E. K., Clavarino, G., Ceppi, M. & Pierre, P. Nat. Methods 6, 275–277 (2009).
Dieterich, D. C. et al. Nat. Neurosci. 13, 897–905 (2010).
tom Dieck, S. et al. Curr. Protoc. Cell Biol. 56, 7.11.1–7.11.29 (2012).
Dörrbaum, A. R., Kochen, L., Langer, J. D. & Schuman, E. M. eLife 7, 489 (2018).
Davis, H. P. & Squire, L. R. Psychol. Bull. 96, 518–559 (1984).
Ho, V. M., Lee, J.-A. & Martin, K. C. Science 334, 623–628 (2011).
Govindarajan, A., Israely, I., Huang, S.-Y. & Tonegawa, S. Neuron 69, 132–146 (2011).
Cajigas, I. J., Will, T. & Schuman, E. M. EMBO J. 29, 2746–2752 (2010).
Aakalu, G., Smith, W. B., Nguyen, N., Jiang, C. & Schuman, E. M. Neuron 30, 489–502 (2001).
Atasoy, D., Aponte, Y., Su, H. H. & Sternson, S. M. J. Neurosci. 28, 7025–7030 (2008).
Woodhead, G. J., Mutch, C. A., Olson, E. C. & Chenn, A. J. Neurosci. 26, 12620–12630 (2006).
Link, A. J., Vink, M. K. S. & Tirrell, D. A. Nat. Protoc. 2, 1879–1883 (2007).
Rangaraju, V., Lauterbach, M. & Schuman, E. M. Cell 176, 73–84 (2019).
Acknowledgements
We thank S. tom Dieck and A. Berken for their assistance in obtaining approval for experiments. We thank N. Fuerst, I. Bartnik and A. Staab for the preparation of cultured hippocampal neurons. We thank M. Lauterbach (MPI for Brain Research) for the MATLAB script used to analyze spine-head volumes. Some plasmids used in this study were gifts from P.C. Shaw (The Chinese University of Hong Kong), C. Hanus (Institute for Psychiatry and Neurosciences of Paris, Inserm U1266) and S. Sternson (Janelia Research Campus; plasmid from Addgene #18925); J.M. Wilson (Perelman School of Medicine, University of Pennylvania; plasmid from Addgene #105558); and A. Chenn (Department of Pathology at the University of Illinois, Chicago; plasmid from Addgene #26646). E.M.S is funded by the Max Planck Society, an Advanced Investigator award from the European Research Council (743216), DFG CRC 1080 and DFG CRC 902. A.B. is supported by an EMBO long-term fellowship (ALTF 331-2017).
Author information
Authors and Affiliations
Contributions
M.H. designed, conducted and analyzed experiments and edited the manuscript. C.G. designed, conducted and analyzed experiments and edited the manuscript. V.R. conducted and analyzed the spine plasticity experiments and edited the manuscript. A.B. conducted experiments and edited the manuscript. E.M.S. designed experiments and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
E.M.S., M.H. and C.G. are employees of the Max Planck Society. A European patent application (application number EP19177622.8) based on the results of this publication has been submitted by the Max Planck Society and the above-mentioned authors (Max Planck Institute of Brain Research). The patent covers the design and proof of concept of a gePSI.
Additional information
Peer review information: Rita Strack was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
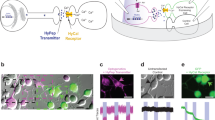
Supplementary Figure 1 gePSI element controls.
a, Scheme of the plasmids used in this study. A plasmid carrying the Tet3G transactivator was co-transfected with plasmids driving the expression of the synthetic ProRIP (1), the gePSI (2) or the separate gePSI chains (3–4) under the control of an inducible pTRE3G or pTRE3G-Bi promoter. b/d, Representative fluorescent images show hippocampal neurons co-transfected with pCMV-Tet3G + pCMV-AcGFP + pTRE3G-Bi-α-Chain (b) or pTRE3G-Bi-β-Chain (d). Nascent protein signal in red, anti-MAP2 immunostaining in blue and GFP in green (white outlines). Scale bar = 20 μm. c/e, Quantification of gePSI – alpha-Chain (c) and – beta-Chain (e) with (4h) and without induction of their expression via dox. Each dot represents the nascent protein signal intensity from one neuron normalized to the mean nascent protein signal of neighboring, untransfected neurons. From left to right: n = 66, 93; mean = 1.11, 0.96; sd = 0.37, 0.48; p-value = 0.1155; Mann-Whitney U (two-tailed) test (c). From left to right: n = 95, 84; mean = 1.13, 0.98; sd = 0.38, 0.46; p-value = 0.1653; Mann-Whitney U (two-tailed) test (e).
Supplementary Figure 2 Detection of gePSI action using FUNCAT and comparison with pharmacological protein synthesis inhibition.
a, Representative fluorescent images of neurons 4h after induction (4h dox) or mock-induction (no dox) of gePSI expression in culture. The translational state of cells was assessed by an alternative metabolic labeling approach using fluorescent non-canonical amino acid tagging (FUNCAT) with the methionine analogue L-Azidohomoalanine (AHA / 4 mM for 1.5h). AHA incorporation was visualized via ClickChemistry (AHA, red), neuron shape is represented with MAP2 (blue) and gePSI-construct bearing neurons additionally express GFP constitutively (green / white outlines). Scale bar = 20 μm. b, Quantification of (a). Each dot represents the nascent protein signal intensity from one neuron normalized to the mean nascent protein signal of neighboring, untransfected neurons. From left to right: n = 39, 44; mean = 1, 0.15; sd = 0.39, 0.07; p-value < 0.0001; Mann-Whitney U (two-tailed) test. c-d, Comparison of gePSI induction effect to anisomycin treatment. c, Representative fluorescent images of hippocampal neurons 4h after induction (4h dox), mock-induction (no dox) or 1h anisomycin treatment. Nascent protein (puromycin, 3 μM for 10 min) in red, anti-MAP2 immunostaining in blue and GFP in green to indicate transfected neurons (white outlines). Scale bar = 20 μm. d, Quantification of (c). Each dot represents the nascent protein signal intensity from one neuron normalized to the mean intensity of the dox minus control. From left to right: n = 23, 32, 31; mean = 1, 0.14, 0.11; sd = 0.28, 0.05, 0.03; p-values: column 1 vs. 2 < 0.0001, column 2 vs. 3 = 0.2032; Kruskal-Wallis test followed by Dunn’s multiple comparison test. The following plasmids were co-transfected in these experiments: pCMV-Tet3G + pTRE3G-Bi-gePSI-pCMV-AcGFP.
Supplementary Figure 3 gePSI expressing cells do not exhibit compromised cell health relative to control cells.
a, Representative images of hippocampal neuron cultures treated without (0h) or with 10 mM H2O2 for 1h or 24h. Cell death was visualized using the propidium iodide (PI) exclusion assay (red) and nuclei were stained with the Hoechst dye (blue). Scale bar = 20 μm. b, Quantification of (a). Each dot represents the PI signal from one neuron normalized to the mean PI intensity of untreated neurons. From left to right: n = 83, 107, 96; mean = 1, 29.80, 67.32; sd = 6.81, 35.43, 19.83; p-values: column 1 vs. 2 < 0.0001, column 2 vs. 3 < 0.0001; Kruskal-Wallis test followed by Dunn’s multiple comparison test. c, Scheme (top) and representative images (bottom) of gePSI- or GFP- (ctrl) transfected primary hippocampal neurons (white outlines) that were continuously induced with 100 ng/ml dox for 1, 2, 4 or 6d (dox was refreshed after 3d). PI signal in red and Hoechst stain in blue. The following plasmids were co-transfected in these experiments: pCMV-Tet3G + pTRE3G-Bi-gePSI-pCMV-AcGFP or pCMV-AcGFP. Scale bar = 20 μm. d, Analysis of (c). Each dot represents the PI signal intensity from one transfected neuron normalized to the mean intensity of the dox minus control. From left to right: n = 46, 38, 36, 39, 32, 46; mean = 1, 1.58, 1.03, 2.32, 5.03, 1.22; sd = 0.56, 1.64, 0.43, 4.23, 12.54, 0.63; p-values: column 1 vs. 2 = 0.7342, column 1 vs. 3 > 0.9999, column 1 vs. 4 = 0.0971, column 1 vs. 5 = 0.0014, column 2 vs. 3 > 0.9999, column 2 vs. 4 > 0.9999, column 3 vs. 4 = 0.8998; Kruskal-Wallis test followed by Dunn’s multiple comparison test.
Supplementary Figure 4 Extended or enhanced gePSI expression in neurons that exhibit retarded protein synthesis recovery.
a, Quantification of nascent protein signal after induction of gePSI transfected hippocampal neurons for 4h using dox concentrations ranging from 1 to 1000 ng/ml. Each dot represents the nascent protein signal intensity from one neuron normalized to the mean nascent protein signal of neighboring, untransfected neurons. In these experiments and those shown below (b-e), the following plasmids were co-transfected: pCMV-Tet3G + pTRE3G-Bi-gePSI-pCMV-AcGFP. From left to right: n = 32, 35, 38, 47, 34; mean = 1.11, 1, 0.27, 0.23, 0.2; sd = 0.3, 0.4, 0.2, 0.11, 0.06; p-values: column 1 vs. 2 > 0.9999, column 1 vs. 3 < 0.0001, column 2 vs. 3 < 0.0001, column 3 vs. 4 > 0.9999, column 3 vs. 5 > 0.9999, column 4 vs. 5 > 0.9999; Kruskal-Wallis test followed by Dunn’s multiple comparison test. b, Representative images of hippocampal neurons transfected with gePSI prior to the addition of dox (no dox) or after a 4h incubation with dox (4h dox), as well as examples of neurons that exhibited recovered protein synthesis levels (recovery-yes) or not (recovery-no) 24h after the dox washout. Nascent protein signal is shown in red, anti-MAP2 immunostaining to visualize neuronal morphology is shown in blue and gePSI mRNA in white. Dashed white outlines indicate transfected neurons. Scale bar = 20 μm. c, Quantification of (b). Each dot represents the mRNA signal from one neuron normalized to the mean mRNA signal of all neurons in the no dox control. From left to right: n = 40, 34, 48, 26; mean = 1, 11.3, 3.4, 0.95; sd = 0.38, 4.56. 1.55, 0.54; p-values: column 1 vs. 2 < 0.0001, column 1 vs. 3 < 0.0001, column 1 vs. 4 > 0.9999, column 2 vs. 3 = 0.0007, column 3 vs. 4 < 0.0001; Kruskal-Wallis test followed by Dunn’s multiple comparison test. d, Representative images showing TUNEL staining in neurons before the induction of gePSI, 4h post induction (4h dox), 24h (4h dox + 24h chase) and 48h (4h dox + 48h chase) after dox washout and positive control. Arrowhead highlights background signal caused by lipofectamine transfection. Scale bar = 5 µm. e, Analysis of (d). TUNEL intensities were normalized to the mean TUNEL intensity of the positive control. From left to right: n = 60, 67, 58, 49, 92; mean = 0.4, 0.32, 0.27, 0.31, 1; sd = 0.12, 0.12, 0.16, 0.13, 0.59; p-values: column 1 vs. 5 < 0.0001, column 2 vs. 5 < 0.0001, column 3 vs. 5 < 0.0001, column 4 vs. 5 < 0.0001; Kruskal-Wallis test followed by Dunn’s multiple comparison test.
Supplementary Figure 5 Cell type-specific protein synthesis inhibition.
a–f, Cell type-specific expression of gePSI in hippocampal cell cultures (containing both neurons and glial cells) or HeLa cells. Specificity was achieved using a Camk2a (a) or a GFAP (b) promoter to restrict transactivator expression in excitatory hippocampal neurons or astrocytes, respectively. The universal mammalian cytomegalovirus (CMV) promoter was used in HeLa cultures (e). Nascent protein signal in red and GFP in green. White outlines indicate transfected cells. The correct cell type for the heterogonous neuronal cultures is verified via FISH (cyan). HeLa cultures were homogenous and representative transfected HeLa cells are visualized with outlines (white dotted line) in the nascent protein-channel. Scale bars = 20 μm. c, d, f, The nascent protein signal in transfected cells was normalized to the respective signal from neighboring, untransfected cells. Each dot represents the mean intensity from one cell. HeLa cells treated with dox are shown relative to the dox minus control. From left to right: n = 21, 20; mean = 1.01, 0.14; sd = 0.26, 0.07; p-value < 0.0001; Mann-Whitney U (two-tailed) test (c). n = 35, 38; mean = 0.89, 0.37; sd = 0.37, 0.36; p-value < 0.0001; Mann-Whitney U (two-tailed) test (d). From left to right: n = 288, 270; mean = 0.84, 0.47; sd = 0.55, 0.36; p-value < 0.0001; Mann-Whitney U (two-tailed) test (f). The following plasmids were co-transfected: pCamk2a-Tet3G or pGFAP-Tet3G or pCMV-Tet3G + pTRE3G-Bi-gePSI-pCamk2a-AcGFP or pTRE3G-Bi-gePSI-pGFAP-AcGFP or pTRE3G-Bi-gePSI-pCMV-AcGFP.
Supplementary Figure 6 Cre-dependent expression of the gePSI.
a, Scheme of the cre-dependent gePSI-plasmid using the cre recombinase for the cell type-specific expression of the gePSI. b, Representative fluorescence images of hippocampal neurons transfected with the floxed Tet3G, and the gePSI with or without Cre recombinase. Nascent protein signal in red, anti-HA immunostaining in “fire” look-up to visualize cre expression and GFP in green (white outlines) to indicate transfected neurons. Scale bar = 20 µm. c, Quantification of (b). Each dot represents the nascent protein signal intensity from one neuron normalized to the mean nascent protein signal of neighboring, untransfected neurons. From left to right: n = 57, 77, 65; mean = 1.13, 1.04, 0.29; sd = 0.32, 0.32, 0.23; p-values: column 1 vs. 2 = 0.5520, column 2 vs. 3 < 0.0001; Kruskal-Wallis test followed by Dunn’s multiple comparison test. d, Representative fluorescence images of hippocampal neurons transfected with the floxed Tet3G, gePSI and Cre recombinase under the Camk2a promoter. Nascent protein signal in red, FISH against Camk2a mRNA in cyan and GFP in green. Scale bar = 20 µm. e, Quantification of (d). Each dot represents the nascent protein signal intensity from one neuron normalized to the mean nascent protein signal of neighboring, untransfected neurons. From left to right: n = 65, 42; mean = 1.1, 0.15; sd = 0.3, 0.04; p-value < 0.0001; f-value = 66.49; two-tailed, unpaired t-test with Welch’s correction (t = 25.41, df = 66.96). The following plasmids were co-transfected: pCAG-Cre-IRES2-eGFP + pCMV-FLEx(inverted)-Tet3g-3xHA-FLEx(inverted) + pTRE3G-Bi-gePSI; pCamk2a-Cre + pCMV-FLEx(inverted)-Tet3g-3xHA-FLEx(inverted) + pTRE3G-Bi-gePSI + pCamk2a-AcGFP.
Supplementary information
Rights and permissions
About this article
Cite this article
Heumüller, M., Glock, C., Rangaraju, V. et al. A genetically encodable cell-type-specific protein synthesis inhibitor. Nat Methods 16, 699–702 (2019). https://doi.org/10.1038/s41592-019-0468-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-019-0468-x