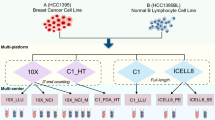
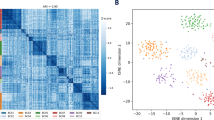
Abstract
Single-cell RNA sequencing is often applied in study designs that include multiple individuals, conditions or tissues. To identify recurrent cell subpopulations in such heterogeneous collections, we developed Conos, an approach that relies on multiple plausible inter-sample mappings to construct a global graph connecting all measured cells. The graph enables identification of recurrent cell clusters and propagation of information between datasets in multi-sample or atlas-scale collections.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
HCA BM and cord blood data were downloaded from the HCA portal (https://preview.data.humancellatlas.org/). The dataset represents a relatively uniform collection of data on well-studied tissues, making it particularly suitable for benchmarking purposes. To reduce calculation times in benchmark evaluations, we took a random subset of the cells from lane 1 of each dataset. By default, 3,000 cells per sample were used (HCA BM + CB 3k datasets). A smaller, 1,000-cell dataset (HCA BM + CB 1k) was used for the more extensive sensitivity analysis (Supplementary Fig. 1f). For Fig. 1i, we combined HCA BM samples with two samples (‘Frozen BMMCs Healthy Donors 1 and 2’) downloaded this from 10x Genomics (https://www.10xgenomics.com/resources/datasets/). This was done to extend the number of samples (x axis in Fig. 1i). The data on breast cancer from Azizi et al.9 were downloaded from GEO (GSE114725) as a count matrix, together with the provided annotations. As shown in the plots (Fig. 2 and Supplementary Fig. 4), the annotations were simplified to collapse patient-specific populations and omit smaller subpopulation distinctions. To demonstrate applicability to different levels of data fragmentation, we reanalyzed the dataset by combining eight individual subjects, 15 subject + tissue combinations or 53 subject + tissue + replicate combinations. The dataset provides a good example of a clinically oriented panel with both tissue- and individual-level heterogeneity. The molecular count data and annotations on lung cancer from Lambrechts et al.12 were downloaded from ArrayExpress (E-MTAB-6149, E-MTAB-6653). The dataset provides an example of a more typical case-control design of a clinically oriented panel. The molecular count data and annotations on non-small-cell lung cancer from Guo et al.11 were downloaded from GEO (GSE99254). The dataset serves as an example of a heterogeneous clinically oriented panel, with limited complexity and numbers of cells in some of the samples. The molecular count data and annotations on head-and-neck cancer from Puram et al.10 were downloaded from GEO (GSE103322). Similar to the data from Guo et al.11, the dataset provides an example of a collection with challenging complexity and cell-number variation in a clinically oriented panel. For the human cortex comparison, the datasets were included as an example of integration of distinct nuclei-based protocols. The count matrix for Hodge et al. (bioRxiv; https://doi.org/10.1101/384826) was downloaded from http://celltypes.brain-map.org/rnaseq. The count matrix from Lake et al. (Nat. Biotechnol. 36, 70–80; 2018) was downloaded from GEO (GSE97930). Tabula Muris mouse data were downloaded from https://tabula-muris.ds.czbiohub.org/. Only cells with at least 1,000 molecules were analyzed. A total of 48 datasets were combined. The mouse cell atlas by Han et al.16 and the relevant annotations were downloaded from http://bis.zju.edu.cn/MCA/. Cell line datasets were excluded. Human pancreas islet data from different platforms, used to demonstrate alignment between different platforms and illustrate mixing controls (Supplementary Fig. 9), were taken from the following sources: 10x Chromium platform data were taken from a publication by Xin et al.19 and downloaded from GEO (GSE114297). Normalized count matrices were used. inDrops platform data were taken from a publication by Baron et al.20 and downloaded from GEO (GSE84133). Only human data (four samples) were used. Normalized count matrices were used. Smart-seq2 platform data were taken from a publication by Segerstolpe et al.21 with count matrices downloaded from ArrayExpress (E-MTAB-5061). Only data from healthy individuals (six samples) were used. For the demonstration of ATAC-seq alignment and alignment between ATAC-seq and RNA-seq (Supplementary Note 2), the following datasets were used: sci-ATAC data from Cusanovich et al.17 were downloaded from the authors’ website (http://atlas.gs.washington.edu/mouse-atac/). Author-provided accessibility scores were used as gene-level input to Conos. sci-CAR data from Cao et al.18 were downloaded from GEO (GSE117089). To increase coverage, the cells were aggregated into groups of ten on the basis of transcriptional similarity (see Supplementary Note 2 for details).
Code availability
Conos is implemented as an R package with C++ optimizations, and is available on GitHub (https://github.com/hms-dbmi/conos) under the GPL-3 open source license. Analysis scripts and intermediate data representations used for the preparation of the manuscript can be found on the author’s website (http://pklab.med.harvard.edu/peterk/conos/).
References
Tabula Muris Consortium. Nature 562, 367–372 (2018).
Regev, A. et al. eLife 6, e27041 (2017).
Hicks, S. C., Townes, F. W., Teng, M. & Irizarry, R. A. Biostatistics 19, 562–578 (2018).
McCarthy, D. J., Campbell, K. R., Lun, A. T. & Wills, Q. F. Bioinformatics 33, 1179–1186 (2017).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Nat. Biotechnol. 36, 411–420 (2018).
Haghverdi, L., Lun, A. T. L., Morgan, M. D. & Marioni, J. C. Nat. Biotechnol. 36, 421–427 (2018).
Neuenschwander, B. E. & Flury, B. D. J. Multivar. Anal. 75, 163–183 (2000).
Ponnapalli, S. P., Saunders, M. A., Van Loan, C. F. & Alter, O. PLoS ONE 6, e28072 (2011).
Azizi, E. et al. Cell 174, 1293–1308 e1236 (2018).
Puram, S. V. et al. Cancer Cell 171, 1611–1624 (2017).
Guo, X. et al. Nat. Med. 24, 978–985 (2018).
Lambrechts, D. et al. Nat. Med. 24, 1277–1289 (2018).
Lun, A. T. L. & Marioni, J. C. Biostatistics 18, 451–464 (2017).
Love, M. I., Huber, W. & Anders, S. Genome Biol. 15, 550 (2014).
Ritchie, M. E. et al. Nucleic Acids Res. 43, e47 (2015).
Han, X. et al. Cell 172, 1091–1107 (2018).
Cusanovich, D. A. et al. Cell 174, 1309–1324 (2018).
Cao, J. et al. Science 361, 1380–1385 (2018).
Xin, Y. et al. Diabetes 67, 1783–1794 (2018).
Baron, M. et al. Cell Syst. 3, 346–360 e344 (2016).
Segerstolpe, A. et al. Cell Metab. 24, 593–607 (2016).
Acknowledgements
N.B. and P.V.K. were supported by the NIH R01HL131768 and NSF-14-532 CAREER awards. D.N. and Y.Z. were supported by the SMTB Alumni Summer Research Program from Zimin Foundation. We would like to thank the HMS Research Computing team for facilitating benchmarking calculations using the O2 cluster.
Author information
Authors and Affiliations
Contributions
N.B., V.P. and P.V.K. implemented the method and ran evaluations. D.N. evaluated label transfer benchmarks and helped with implementation. Y.L. implemented the interactive view of hierarchical communities. S.D. and K.K. applied the method to integration of neuronal atlases. P.V.K. designed and oversaw the study and drafted the manuscript with help from V.P.
Corresponding author
Ethics declarations
Competing interests
P.V.K. serves on the Scientific Advisory Board to Celsius Therapeutics Inc.
Additional information
Peer Review Information: Nicole Rusk was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–19 and Supplementary Notes 1 and 2
Rights and permissions
About this article
Cite this article
Barkas, N., Petukhov, V., Nikolaeva, D. et al. Joint analysis of heterogeneous single-cell RNA-seq dataset collections. Nat Methods 16, 695–698 (2019). https://doi.org/10.1038/s41592-019-0466-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-019-0466-z
This article is cited by
-
Artificial intelligence and illusions of understanding in scientific research
Nature (2024)
-
Dictionary learning for integrative, multimodal and scalable single-cell analysis
Nature Biotechnology (2024)
-
Single-cell analysis of immune and stroma cell remodeling in clear cell renal cell carcinoma primary tumors and bone metastatic lesions
Genome Medicine (2024)
-
GoM DE: interpreting structure in sequence count data with differential expression analysis allowing for grades of membership
Genome Biology (2023)
-
MCProj: metacell projection for interpretable and quantitative use of transcriptional atlases
Genome Biology (2023)