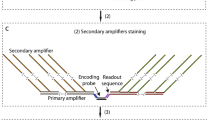
Abstract
Fluorescence in situ hybridization (FISH) reveals the abundance and positioning of nucleic acid sequences in fixed samples. Despite recent advances in multiplexed amplification of FISH signals, it remains challenging to achieve high levels of simultaneous amplification and sequential detection with high sampling efficiency and simple workflows. Here we introduce signal amplification by exchange reaction (SABER), which endows oligonucleotide-based FISH probes with long, single-stranded DNA concatemers that aggregate a multitude of short complementary fluorescent imager strands. We show that SABER amplified RNA and DNA FISH signals (5- to 450-fold) in fixed cells and tissues. We also applied 17 orthogonal amplifiers against chromosomal targets simultaneously and detected mRNAs with high efficiency. We then used 10-plex SABER-FISH to identify in vivo introduced enhancers with cell-type-specific activity in the mouse retina. SABER represents a simple and versatile molecular toolkit for rapid and cost-effective multiplexed imaging of nucleic acid targets.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
All raw and processed data are available from the authors upon reasonable request.
Code availability
The complete set of CellProfiler38,60 pipelines used and example input images for each are available at https://github.com/brianbeliveau/SABER. PD3D, a package of MATLAB functions for detecting SABER puncta (or other fluorescent puncta) in 3D and assigning puncta to cells in a watershed segmentation, is available at https://github.com/ewest11/PD3D. Functions used for image processing are available at http://saber.fish or http://saber-fish.net/.
References
Pardue, M. L. & Gall, J. G. Molecular hybridization of radioactive DNA to the DNA of cytological preparations. Proc. Natl Acad. Sci. USA 64, 600–604 (1969).
Riegel, M. Human molecular cytogenetics: from cells to nucleotides. Genet. Mol. Biol. 37, 194–209 (2014).
Bolzer, A. et al. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol. 3, 0826–0842 (2005).
Femino, A. M., Fay, F. S., Fogarty, K. & Singer, R. H. Visualization of single RNA transcripts in situ. Science 280, 585–590 (1998).
Raj, A., van den Bogaard, P., Rifkin, S. A., van Oudenaarden, A. & Tyagi, S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods 5, 877–879 (2008).
Schröck, E. et al. Multicolor spectral karyotyping of human chromosomes. Science 273, 494–497 (1996).
Lubeck, E. & Cai, L. Single-cell systems biology by super-resolution imaging and combinatorial labeling. Nat. Methods 9, 743–748 (2012).
Jungmann, R. et al. Multiplexed 3D cellular super-resolution imaging with DNA-PAINT and Exchange-PAINT. Nat. Methods 11, 313–318 (2014).
Schueder, F. et al. Universal super-resolution multiplexing by DNA exchange. Angew. Chem. Int. Ed. Engl. 56, 4052–4055 (2017).
Wang, Y. et al. Rapid sequential in situ multiplexing with DNA exchange imaging in neuronal cells and tissues. Nano Lett. 17, 6131–6139 (2017).
Wang, S. et al. Spatial organization of chromatin domains and compartments in single chromosomes. Science 353, 598–602 (2016).
Bintu, B. et al. Super-resolution chromatin tracing reveals domains and cooperative interactions in single cells. Science 362, eaau1783 (2018).
Codeluppi, S. et al. Spatial organization of the somatosensory cortex revealed by osmFISH. Nat. Methods 15, 932–935 (2018).
Lubeck, E., Coskun, A. F., Zhiyentayev, T., Ahmad, M. & Cai, L. Single-cell in situ RNA profiling by sequential hybridization. Nat. Methods 11, 360–361 (2014).
Chen, K. H., Boettiger, A. N., Moffitt, J. R., Wang, S. & Zhuang, X. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090 (2015).
Levesque, M. J. & Raj, A. Single-chromosome transcriptional profiling reveals chromosomal gene expression regulation. Nat. Methods 10, 246–248 (2013).
Shah, S. et al. Dynamics and spatial genomics of the nascent transcriptome by intron seqFISH. Cell 174, 363–376 (2018).
Kerstens, H. M., Poddighe, P. J. & Hanselaar, A. G. A novel in situ hybridization signal amplification method based on the deposition of biotinylated tyramine. J. Histochem. Cytochem. 43, 347–352 (1995).
Player, A. N., Shen, S. P., Kenny, D., Antao, V. P. & Kolberg, J. A. Single-copy gene detection using branched DNA (bDNA) in situ hybridization. J. Histochem. Cytochem. 49, 603–611 (2001).
Wang, F. et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J. Mol. Diagn. 14, 22–29 (2012).
Beliveau, B. J. et al. Single-molecule super-resolution imaging of chromosomes and in situ haplotype visualization using Oligopaint FISH probes. Nat. Commun. 6, 7147 (2015).
Lizardi, P. et al. Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nat. Genet. 19, 225–232 (1998).
Dirks, R. M. & Pierce, N. A. Triggered amplification by hybridization chain reaction. Proc. Natl Acad. Sci. USA 101, 15275–15278 (2004).
Choi, H. M. T. et al. Programmable in situ amplification for multiplexed imaging of mRNA expression. Nat. Biotechnol. 28, 1208–1212 (2010).
Choi, H. M., Beck, V. A. & Pierce, N. A. Next-generation in situ hybridization chain reaction: higher gain, lower cost, greater durability. ACS Nano 8, 4284–4294 (2014).
Shah, S. et al. Single-molecule RNA detection at depth via hybridization chain reaction and tissue hydrogel embedding and clearing. Development 92, 2862–2867 (2016).
Rouhanifard, S. H. et al. ClampFISH detects individual nucleic acid molecules using click chemistry–based amplification. Nat. Biotechnol. 37, 84–89 (2018).
Nagendran, M., Riordan, D. P., Harbury, P. B. & Desai, T. J. Automated cell-type classification in intact tissues by single-cell molecular profiling. eLife 7, e30510 (2018).
Wang, X. et al. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science 361, eaat5691 (2018).
Kishi, J. Y., Schaus, T. E., Gopalkrishnan, N., Xuan, F. & Yin, P. Programmable autonomous synthesis of single-stranded DNA. Nat. Chem. 10, 155–164 (2018).
Beliveau, B. J. et al. Versatile design and synthesis platform for visualizing genomes with Oligopaint FISH probes. Proc. Natl Acad. Sci. USA 109, 21301–21306 (2012).
Lee, C. S., Davis, R. W. & Davidson, N. A physical study by electron microscopy of the terminally repetitious, circularly permuted DNA from the coliphage particles of Escherichia coli 15. J. Mol. Biol. 48, 1–22 (1970).
Beliveau, J. et al. OligoMiner provides a rapid, flexible environment for the design of genome-scale oligonucleotide in situ hybridization probes. Proc. Natl Acad. Sci. USA 115, E2183–E2192 (2018).
Xu, Q., Schlabach, M. R., Hannon, G. J. & Elledge, S. J. Design of 240,000 orthogonal 25mer DNA barcode probes. Proc. Natl Acad. Sci. USA 106, 2289–2294 (2009).
Dirks, R. M. & Pierce, N. A. A partition function algorithm for nucleic acid secondary structure including pseudoknots. J. Comput. Chem. 24, 1664–1677 (2003).
Dirks, R. M. & Pierce, N. A. An algorithm for computing nucleic acid base-pairing probabilities including pseudoknots. J. Comput. Chem. 25, 1295–1304 (2004).
Dirks, R. M., Bois, J. S., Schaeffer, J. M., Winfree, E. & Pierce, N. A. Thermodynamic analysis of interacting nucleic acid strands. SIAM Rev. 49, 65–88 (2007).
Carpenter, A. E. et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7, R100 (2006).
Macosko, E. Z. et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015).
Shekhar, K. et al. Comprehensive classification of retinal bipolar neurons by single-cell transcriptomics. Cell 166, 1308–1323 (2016).
Mosaliganti, K. R., Noche, R. R., Xiong, F., Swinburne, I. A. & Megason, S. G. ACME: automated cell morphology extractor for comprehensive reconstruction of cell membranes. PLoS Comput. Biol. 8, e1002780 (2012).
Solovei, I. et al. Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell 137, 356–368 (2009).
Shah, S., Lubeck, E., Zhou, W. & Cai, L. In situ transcription profiling of single cells reveals spatial organization of cells in the mouse hippocampus. Neuron 92, 342–357 (2016).
Emerson, M. M. & Cepko, C. L. Identification of a retina-specific Otx2 enhancer element active in immature developing photoreceptors. Dev. Biol. 360, 241–255 (2011).
ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Matsuda, T. & Cepko, C. L. Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc. Natl Acad. Sci. USA 101, 16–22 (2004).
Saka, S. K. et al. Highly multiplexed in situ protein imaging with signal amplification by Immuno-SABER. Nat. Biotechnol. (in the press).
Frieda, K. L. et al. Synthetic recording and in situ readout of lineage information in single cells. Nature 541, 107–111 (2017).
Yildirim, E., Sadreyev, R. I., Pinter, S. F. & Lee, J. T. X-chromosome hyperactivation in mammals via nonlinear relationships between chromatin states and transcription. Nat. Struct. Mol. Biol. 19, 56–61 (2011).
Kent, W. J. et al. The Human Genome Browser at UCSC. Genome Res. 12, 996–1006 (2002).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Marçais, G. & Kingsford, C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27, 764–770 (2011).
Casanova, M. et al. Heterochromatin reorganization during early mouse development requires a single-stranded noncoding transcript. Cell Rep. 4, 1156–1167 (2013).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Beliveau, B. J., Apostolopoulos, N. & Wu, C. Visualizing genomes with Oligopaint FISH probes. Curr. Protoc. Mol. Biol. 2014, 14.23.1–14.23.20 (2014).
Ran, F. A. et al. Genome engineering using the CRISPR–Cas9 system. Nat. Protoc. 8, 2281–2308 (2013).
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
McQuin, C. et al. CellProfiler 3.0: next-generation image processing for biology. PLoS Biol. 16, 1–17 (2018).
Linkert, M. et al. Metadata matters: access to image data in the real world. J. Cell Biol. 189, 777–782 (2010).
Yushkevich, P. A. et al. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage 31, 1116–1128 (2006).
Marr, D. & Hildreth, E. Theory of edge detection. Proc. R. Soc. Lond. B Biol. Sci. 207, 187–217 (1980).
Plaisier, S., Taschereau, R., Wong, J. & Graeber, T. Rank-rank hypergeometric overlap: identification of statistically significant overlap between gene-expression signatures. Nucleic Acids Res. 38, e169 (2010).
Hunter, J. D. Matplotlib: a 2D graphics environment. Comput. Sci. Eng. 9, 90–95 (2007).
Waskom, M. et al. mwaskom/seaborn: v0.8.1. https://doi.org/10.5281/zenodo.883859 (2017).
Oliphant, T. E. A Guide to NumPy (Trelgol Publishing, 2006).
McKinney, W. Data structures for statistical computing in Python. in Proc. 9th Python in Science Conference (eds. van der Walt, S. & Millman, J.) 51–56 (SciPy, 2010).
Cock, P. J. A. et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25, 1422–1423 (2009).
Kassambara, A. ggpubr: ‘ggplot2’ based publication ready plots, version 0.1.7. https://cran.r-project.org/web/packages/ggpubr/index.html (2018).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer, 2016).
R Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2013).
Acknowledgements
The authors thank B. Fields, S. Kennedy, J.A. Abed, T. Wu, T. Ferrante, N. Liu, F. Dannenberg, M. Cicconet, P.M. Llopis and the Microscopy Resources on the North Quad (MicRoN) at Harvard Medical School for discussions and technical support. We also thank the ENCODE consortium and J. Stamatoyannopoulos (University of Washington) for retina DHS data. This work was supported by the National Institutes of Health (under grants 1R01EB018659-01 to P.Y., 1UG3HL145600 to P.Y., 1R01GM124401 to P.Y., 1U01MH106011-01 to P.Y., 1DP1GM133052 to P.Y., 5K99EY028215-02 to S.W.L. and a T32 training grant GM096911 supporting E.R.W.), the Office of Naval Research (under grants N00014-16-1-2410 to P.Y. and N00014-18-1-2549 to P.Y.), the National Science Foundation (under grant CCF-1317291 to P.Y. and a Graduate Research Fellowship to J.Y.K.), the Howard Hughes Medical Institute (C.L.C.), the Damon Runyon Cancer Research Foundation (under a fellowship to B.J.B.), the Uehara Memorial Foundation (under a fellowship to H.M.S.), the Human Frontier Science Program (under fellowship LT000048/2016-L to S.K.S.), EMBO (under fellowship ALTF 1278-2015 to S.K.S.) and the Wyss Institute’s Molecular Robotics Initiative (MRI) (P.Y., J.Y.K. and B.J.B.).
Author information
Authors and Affiliations
Contributions
J.Y.K., S.W.L., B.J.B., E.R.W., C.L.C. and P.Y. conceived the study. J.Y.K. and B.J.B. designed SABER probes, designed and executed cell experiments and analyzed cell data. S.W.L. designed and executed tissue experiments. E.R.W. developed the analytical pipeline and methods for tissue cell segmentation and puncta quantification. J.Y.K., S.W.L., B.J.B., E.R.W., A.Z., S.K.S., H.M.S. and Y.W. contributed to optimizing and performing experimental protocols and obtaining data. J.Y.K., S.W.L., B.J.B., E.R.W., C.L.C. and P.Y. wrote the manuscript. All authors edited and approved the manuscript. C.L.C. and P.Y. supervised the work.
Corresponding authors
Ethics declarations
Competing interests
A provisional US patent has been filed based on this work (PCT/US2018/013019). P.Y. is cofounder of Ultivue, Inc. and NuProbe Global.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–10 and Supplementary Note
Supplementary Protocols
Step-by-step instructions for designing and applying SABER-FISH
Supplementary Table 1
SABER sequences. PER, imager, and probe pool sequences.
Supplementary Table 2
SABER simulated parameters. Melting temperatures of PER, probe and bridge sequences under different formamide conditions, as well as cross-talk probabilities.
Supplementary Table 3
SABER experimental conditions. Detailed PER, ISH and fluorescent-hybridization conditions for each experiment.
Supplementary Table 4
SABER puncta counts. Cell counts, puncta counts, puncta sizes, signal-to-noise values, cross-correlation values and biological replicate information for experiments.
Rights and permissions
About this article
Cite this article
Kishi, J.Y., Lapan, S.W., Beliveau, B.J. et al. SABER amplifies FISH: enhanced multiplexed imaging of RNA and DNA in cells and tissues. Nat Methods 16, 533–544 (2019). https://doi.org/10.1038/s41592-019-0404-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-019-0404-0
This article is cited by
-
Tigerfish designs oligonucleotide-based in situ hybridization probes targeting intervals of highly repetitive DNA at the scale of genomes
Nature Communications (2024)
-
Thermal-plex: fluidic-free, rapid sequential multiplexed imaging with DNA-encoded thermal channels
Nature Methods (2024)
-
Highly sensitive spatial transcriptomics using FISHnCHIPs of multiple co-expressed genes
Nature Communications (2024)
-
DNA-barcoded signal amplification for imaging mass cytometry enables sensitive and highly multiplexed tissue imaging
Nature Methods (2023)
-
SGF29 nuclear condensates reinforce cellular aging
Cell Discovery (2023)