Abstract
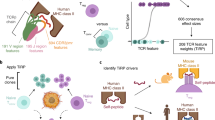
T cell receptor (TCR) ligand discovery is essential for understanding and manipulating immune responses to tumors. We developed a cell-based selection platform for TCR ligand discovery that exploits a membrane transfer phenomenon called trogocytosis. We discovered that T cell membrane proteins are transferred specifically to target cells that present cognate peptide–major histocompatibility complex (MHC) molecules. Co-incubation of T cells expressing an orphan TCR with target cells collectively presenting a library of peptide–MHCs led to specific labeling of cognate target cells, enabling isolation of these target cells and sequencing of the cognate TCR ligand. We validated this method for two clinically employed TCRs and further used the platform to identify the cognate neoepitope for a subject-derived neoantigen-specific TCR. Thus, target cell trogocytosis is a robust tool for TCR ligand discovery that will be useful for studying basic tumor immunology and identifying new targets for immunotherapy.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The original NGS DNA-seq data have been deposited in the Sequence Read Archive under accession numbers SRR8217181, SRR8217182 and SRR8217183. The data that support the findings of this study are available from the corresponding author upon request. Source data for Figs. 2, 4 and 5 are available online.
References
Dembić, Z. et al. Transfer of specificity by murine alpha and beta T-cell receptor genes. Nature 320, 232–238 (1986).
Schumacher, T. N. & Schreiber, R. D. Neoantigens in cancer immunotherapy. Science 348, 69–74 (2015).
Sollid, L. M. et al. Small bowel, celiac disease and adaptive immunity. Dig. Dis. 33, 115–121 (2015).
Rosenberg, S. A. & Restifo, N. P. Adoptive cell transfer as personalized immunotherapy for human cancer. Science 348, 62–68 (2015).
Kontos, S., Grimm, A. J. & Hubbell, J. A. Engineering antigen-specific immunological tolerance. Curr. Opin. Immunol. 35, 80–88 (2015).
Dolton, G. et al. More tricks with tetramers: a practical guide to staining T cells with peptide-MHC multimers. Immunology 146, 11–22 (2015).
Bentzen, A. K. et al. Large-scale detection of antigen-specific T cells using peptide-MHC-I multimers labeled with DNA barcodes. Nat. Biotechnol. 34, 1037–1045 (2016).
Gros, A. et al. Prospective identification of neoantigen-specific lymphocytes in the peripheral blood of melanoma patients. Nat. Med. 22, 433–438 (2016).
Strønen, E. et al. Targeting of cancer neoantigens with donor-derived T cell receptor repertoires. Science 352, 1337–1341 (2016).
Briggs, A. W. et al. Tumor-infiltrating immune repertoires captured by single-cell barcoding in emulsion. bioRxiv Preprint at https://www.biorxiv.org/content/early/2017/05/05/134841 (2017).
Duhen, T. et al. Co-expression of CD39 and CD103 identifies tumor-reactive CD8 T cells in human solid tumors. Nat. Commun. 9, 2724 (2018).
Gros, A. et al. PD-1 identifies the patient-specific CD8+ tumor-reactive repertoire infiltrating human tumors. J. Clin. Invest. 124, 2246–2259 (2014).
Han, A., Glanville, J., Hansmann, L. & Davis, M. M. Linking T-cell receptor sequence to functional phenotype at the single-cell level. Nat. Biotechnol. 32, 684–692 (2014).
Linnemann, C., Mezzadra, R. & Schumacher, T. N. TCR repertoires of intratumoral T-cell subsets. Immunol. Rev. 257, 72–82 (2014).
Pasetto, A. et al. Tumor- and neoantigen-reactive T-cell receptors can be identified based on their frequency in fresh tumor. Cancer Immunol. Res. 4, 734–743 (2016).
Prickett, T. D. et al. Durable complete response from metastatic melanoma after transfer of autologous T cells recognizing 10 mutated tumor antigens. Cancer Immunol. Res. 4, 669–678 (2016).
Simoni, Y. et al. Bystander CD8+ T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature 557, 575–579 (2018).
Joly, E. & Hudrisier, D. What is trogocytosis and what is its purpose? Nat. Immunol. 4, 815 (2003).
Daubeuf, S., Lindorfer, M. A., Taylor, R. P., Joly, E. & Hudrisier, D. The direction of plasma membrane exchange between lymphocytes and accessory cells by trogocytosis is influenced by the nature of the accessory cell. J. Immunol. 184, 1897–1908 (2010).
Cone, R. E., Sprent, J. & Marchalonis, J. J. Antigen-binding specificity of isolated cell-surface immunoglobulin from thymus cells activated to histocompatibility antigens. Proc. Natl Acad. Sci. USA 69, 2556–2560 (1972).
Huang, J. F. et al. TCR-mediated internalization of peptide-MHC complexes acquired by T cells. Science 286, 952–954 (1999).
Hwang, I. et al. T cells can use either T cell receptor or CD28 receptors to absorb and internalize cell surface molecules derived from antigen-presenting cells. J. Exp. Med. 191, 1137–1148 (2000).
Uzana, R. et al. Human T cell crosstalk is induced by tumor membrane transfer. PLoS ONE 10, e0118244 (2015).
He, T. et al. Bidirectional membrane molecule transfer between dendritic and T cells. Biochem. Biophys. Res. Commun. 359, 202–208 (2007).
Kim, H. R. et al. T cell microvilli constitute immunological synaptosomes that carry messages to antigen-presenting cells. Nat. Commun. 9, 3630 (2018).
Bethune, M. T., Comin-Anduix, B., Hwang Fu, Y. H., Ribas, A. & Baltimore, D. Preparation of peptide-MHC and T-cell receptor dextramers by biotinylated dextran doping. Biotechniques 62, 123–130 (2017).
Bethune, M. T. et al. Isolation and characterization of NY-ESO-1-specific T cell receptors restricted on various MHC molecules. Proc. Natl Acad. Sci. USA 115, E10702–E10711 (2018).
Skipper, J. C. et al. Mass-spectrometric evaluation of HLA-A*0201-associated peptides identifies dominant naturally processed forms of CTL epitopes from MART-1 and gp100. Int. J. Cancer 82, 669–677 (1999).
Wooldridge, L. et al. Interaction between the CD8 coreceptor and major histocompatibility complex class I stabilizes T cell receptor-antigen complexes at the cell surface. J. Biol. Chem. 280, 27491–27501 (2005).
Bassani-Sternberg, M. & Coukos, G. Mass spectrometry-based antigen discovery for cancer immunotherapy. Curr. Opin. Immunol. 41, 9–17 (2016).
Bethune, M. T. & Joglekar, A. V. Personalized T cell-mediated cancer immunotherapy: progress and challenges. Curr. Opin. Biotechnol. 48, 142–152 (2017).
Truscott, S. M. et al. Disulfide bond engineering to trap peptides in the MHC class I binding groove. J. Immunol. 178, 6280–6289 (2007).
Joglekar, A. V. et al. T cell antigen discovery via signaling and antigen-presenting bifunctional receptors. Nat. Methods https://doi.org/10.1038/s41592-018-0304-8 (2019).
Gee, M. H. et al. Antigen identification for orphan T cell receptors expressed on tumor-infiltrating lymphocytes. Cell 172, 549–563 (2018).
González-Galarza, F. F. et al. Allele frequency net 2015 update: new features for HLA epitopes, KIR and disease and HLA adverse drug reaction associations. Nucleic Acids Res. 43, D784–D788 (2015).
Tran, E. et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science 344, 641–645 (2014).
Lu, Y. C. & Robbins, P. F. Cancer immunotherapy targeting neoantigens. Semin. Immunol. 28, 22–27 (2016).
Morgan, R. A. et al. Cancer regression and neurological toxicity following anti-MAGE-A3 TCR gene therapy. J. Immunother. 36, 133–151 (2013).
Li, G., Wong, S., Bethune, T. M. & Baltimore, D. Trogocytosis-based cell platform for TCR ligand discovery. Protocol Exchange https://doi.org/10.1038/protex.2018.127 (2019).
Bethune, M. et al. Domain-swapped T cell receptors improve the safety of TCR gene therapy. eLife 5, e19095 (2016).
Neri, S., Mariani, E., Meneghetti, A., Cattini, L. & Facchini, A. Calcein-acetyoxymethyl cytotoxicity assay: standardization of a method allowing additional analyses on recovered effector cells and supernatants. Clin. Diagn. Lab. Immunol. 8, 1131–1135 (2001).
Acknowledgements
We thank I. Antoshechkin (Millard and Muriel Jacobs Genetics and Genomics Laboratory, Caltech) for deep DNA sequencing, and D. Perez, J. Tijerina and R.A. Diamond (Flow Cytometry Facility, Caltech) for cell sorting. This work was supported by the Prostate Cancer Foundation Challenge Award 15CHAL02 to D.B., O.N.W., L.Y. and M.T.B., and the National Cancer Institute (grant 1U54 CA199090-01 to J.R.H.). M.T.B. is the recipient of a Jane Coffin Childs Postdoctoral Fellowship. A.R. was supported by National Institutes of Health (NIH) grant R35 CA197633. G.L. was supported by the Parker Institute for Cancer Immunotherapy. J.T.K. was supported by NIH/National Center for Advancing Translational Science UCLA CTSI grant KL2TR001882.
Author information
Authors and Affiliations
Contributions
M.T.B. conceived of the approach. G.L. and M.T.B. designed research. G.L., M.T.B., S.W., A.V.J., M.T.L., J.K.W., J.T.K., Y.S., Y.L. and D.C. performed experiments. S.P., J.M.Z., A.R. and J.R.H. provided critical reagents and analyzed results. O.N.W. analyzed results. G.L., M.T.B. and D.B. analyzed results and wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
M.T.B., G.L., J.T.K., S.W. and D.B. are co-inventors on a patent application concerning the described technology, which is licensed to PACT Pharma, Inc. J.R.H. and A.R. are directors and consultants of PACT; D.B. is a consultant of PACT and head of their scientific advisory board; M.T.B. and S.P. are employees of PACT; J.M.Z. is a consultant of PACT; and each of the foregoing individuals has equity interests in PACT.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Establishment of Jurkat cells expressing F5 TCR or 1G4 TCR and K562 cells expressing single-chain trimer (SCT) of HLA-A2/MART126-35(A27L) or HLA-A2/NYESO1157-165(C165V).
(a) A retroviral vector co-delivered F5 TCR or 1G4 TCR genes carrying either human or murine TCR constant regions together with LNGFRΔ, a transduction marker comprising low-affinity nerve growth factor receptor with the intracellular domain truncated, to Jurkat cells. A lentiviral vector co-delivered an SCT containing MART1 or NYESO1 peptide with ZsGreen as a transduction marker to K562 cells. SCTs are composed of a single polypeptide chain with a linear composition of antigenic peptide, β2-microglobulin, and HLA-A2 domains via flexible glycine-serine linkers. (b) Resolution via flow cytometry of Jurkat and K562 cells. The Jurkat T cells and K562 cells were resolved by gating on the LNGFR+ population and the ZsGreen+ population, respectively.
Supplementary Figure 2 Target cell trogocytosis occurs in an antigen-specific manner.
(a) ZsGreen cannot be transferred from K562 cells to Jurkat cells. Representative flow cytometry plot of Zsgreen level in F5-CD8-Jurkat or 1G4-CD8-Jurkat following co-incubation with MART1-K562. (b,c)The kinetics of trogocytosis for TCR–cognate pMHC interaction. Co-incubation of 1G4-CD8-Jurkat cells with NYESO1-K562 or F5-CD8-Jurkat cells with MART1-K562 for indicated time and trogocytosis was assessed using anti-muTCR antibody (n = 3). Data are presented as mean ± s.e.m. (d) Immunofluorescence staining of co-incubated F5-CD8-Jurkat with NYESO-K562 (Zsgreen+) or MART1-K562 (Zsgreen+) by MART1 pMHC dextramer (yellow). Data in this figure are representative of at least two independent experiments.
Supplementary Figure 3 Trogocytosis can be tracked by multiple protein transfer.
(a) Antigen-specific transfer of the T cell membrane proteins, CD3 and CD8, from CD8-expressing F5-Jurkat and 1G4-Jurkat cells to K562 cells (ZsGreen+), as assessed by anti-CD3 and anti-CD8 antibodies (5:1 J:K). (b) Antigen-specific transfer of the K562 cell membrane protein, HLA-A2, from K562 cells to Jurkat cells (ZsGreen–), as assessed by an anti-HLA-A2 antibody (5:1 J:K). (c, d) Concomitant reduction of membrane proteins from donor cells. Data are representative of three independent experiments.
Supplementary Figure 4 Trogocytosis occurs among various TCR and pMHC allele pairs.
(a) Antigen-specific trogocytosis between Jurkat expressing B07-, B18- or C03-restricted NY-ESO-1-specific TCRs and K562 expressing their cognate antigens (10:1 J:K). (b) Comparison of trogocytosis capability 1G4-Jurkat cells or Jurkat cell expressing four other novel A2-restricted NYESO-specific TCRs with NYESO1-K562 cells (10:1 J:K). (c) Comparison of trogocytosis capability F5-Jurkat cells or Jurkat cell expressing low-affinity M1W-TCR with MART1-K562 cells (5:1 J:K). Data are presented as mean ± s.e.m. and are representative of two independent experiments.
Supplementary Figure 5 Antigen-specific TCR transfer occurs from donor cells to acceptor cells regardless of cell identity.
(a) Antigen-specific transfer of TCR after co-incubation of a 5:1 ratio of F5 or 1G4 TCR-K562 cells and MART1 or NYESO1 SCT-Jurkat cells (ZsGreen+) as assessed by an anti-muTCR antibody. (b) Antigen-specific transfer of TCR after same-cell-type co-incubation of a 5:1 ratio of F5 or 1G4 TCR-K562 and MART1 or NYESO1 SCT-K562 (ZsGreen+) as assessed by an anti-muTCR antibody. (c) Antigen-specific transfer of TCR after co-incubation of a 1:1 ratio of F5 or 1G4 TCR-Jurkat and MART1 or NYESO1 SCT-Jurkat (ZsGreen+) as assessed by an anti-muTCR antibody. Mean and s.e.m. for each group is shown (n = 3). Data are representative of two independent experiments.
Supplementary Figure 6 Histographic visualization of trogocytosis capability based on peptide dosing and variants.
(a) Comparison of trogocytosis capability of murinized F5-Jurkat cells with A2-K562 cells loaded with different doses of cognate MART1 heteroclitic peptide (ELAGIGILTV). (b) Comparison of trogocytosis capability of murinized 1G4-Jurkat cells with A2-K562 cells loaded with different doses of cognate NYESO1 heteroclitic peptide (SLLMWITQV). (c) Comparison of trogocytosis capability of murinized F5-Jurkat cells with A2-K562 cells loaded with same dose of different MART1 peptide variants. (d) Comparison of trogocytosis capability of murinized 1G4-Jurkat cells with A2-K562 cells loaded with same dose of different NYESO1 peptide. Data are representative of two independent experiments.
Supplementary Figure 7 Target cell trogocytosis is enhanced by coexpression of CD8.
(a) Comparison of trogocytosis capability of CD8+ or CD8– murinized F5-Jurkat cells with MART1-K562 or NYESO1-K562 cells. (b) Comparison of trogocytosis capability of CD8+ or CD8– murinized 1G4-Jurkat cells with MART1-K562 or NYESO1-K562 cells. (c) Comparison of trogocytosis capability of CD8+ or CD8– M1W-Jurkat cells with MART1-K562 cells. Data are representative of two independent experiments.
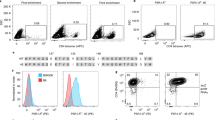
Supplementary Figure 8 Target cell trogocytosis resolves cognate antigen-expressing target cells from noncognate antigen-expressing cells.
(a) Schematic of experiment. (b) Representative flow cytometry plots for a 1:1 mixture of NYESO1-K562 and MART1-K562 cells co-incubated with either 1G4-Jurkat (top) or F5-Jurkat (bottom) cells (10:1:1 Jurkat: NYESO1-K562:MART1-K562). The trogocytosis+ and trogocytosis– populations were verified for antigen specificity by use of either F5 TCR or 1G4 TCR dextramer staining. Data are representative of two independent experiments. (c) Quantification of antigen-specific target cell in trogocytosis+ and trogocytosis– populations via F5 TCR or 1G4 TCR dextramer staining (n = 3). Statistical analysis of quantification was performed using unpaired two-tailed Student’s t-test. Data in b, c are presented as mean ± s.e.m. and are representative of two independent experiments.
Supplementary Figure 9 Flow cytometry plot for the first- and second-round sortings from co-incubation.
(a,b,c) F5-Jurkat, 1G4-Jurkat or neoTCR-Jurkat cells (E2-Crimson+CD8+) were co-incubated with K562 library cells and trogocytosis+ A2-SCT-K562 cells (E2-Crimson–eGFP+TCR+) in red box were gated for sorting using FACS. Data are representative of two or three independent experiments.
Supplementary Figure 10 Comparison of trogocytosis capacity of NeoTCR-CD8-Jurkat cells with MART1-K562, USP7mut-K562 or USP7wt-K562 cells.
The predicted affinity of mutUSP7 and wtUSP7 for the HLA-A2 binding groove by NetMHC are 33 nM and 7 μM, respectively. Data are presented as mean ± s.e.m. (n = 3) and are representative of two independent experiments.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–10 and Supplementary Note 1.
Supplementary Protocol
Reagents
Supplementary Table 1
Epitopes in the A-2 restricted SCT cDNA library.
Supplementary Table 2
Epitopes in the neoantigen SCT cDNA library.
Rights and permissions
About this article
Cite this article
Li, G., Bethune, M.T., Wong, S. et al. T cell antigen discovery via trogocytosis. Nat Methods 16, 183–190 (2019). https://doi.org/10.1038/s41592-018-0305-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-018-0305-7
This article is cited by
-
Novel insights into TCR-T cell therapy in solid neoplasms: optimizing adoptive immunotherapy
Experimental Hematology & Oncology (2024)
-
Neoantigen-targeted TCR-engineered T cell immunotherapy: current advances and challenges
Biomarker Research (2023)
-
The screening, identification, design and clinical application of tumor-specific neoantigens for TCR-T cells
Molecular Cancer (2023)
-
CTLA-4 tail fusion enhances CAR-T antitumor immunity
Nature Immunology (2023)
-
Selenium-based metabolic oligosaccharide engineering strategy for quantitative glycan detection
Nature Communications (2023)