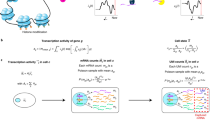
Abstract
Single-cell RNA-seq makes it possible to characterize the transcriptomes of cell types across different conditions and to identify their transcriptional signatures via differential analysis. Our method detects changes in transcript dynamics and in overall gene abundance in large numbers of cells to determine differential expression. When applied to transcript compatibility counts obtained via pseudoalignment, our approach provides a quantification-free analysis of 3′ single-cell RNA-seq that can identify previously undetectable marker genes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Code availability
The code required to conduct the simulations and reproduce the analyses is available at https://github.com/pachterlab/NYMP_2018. We also have provided the Github repository that was zipped at the time of manuscript acceptance as Supplementary Software.
References
Soneson, C. & Robinson, M. D. Nat. Methods 15, 255–261 (2018).
Kharchenko, P. V., Silberstein, L. & Scadden, D. T. Nat. Methods 11, 740–742 (2014).
Finak, G. et al. Genome Biol. 16, 278 (2015).
Yamazaki, T. et al. Genes Dev. 32, 1161–1174 (2018).
Vitting-Seerup, K. & Sandelin, A. Mol. Cancer Res. 15, 1206–1220 (2017).
Arzalluz-Luque, Á. & Conesa, A. Genome Biol. 19, 110 (2018).
Gupta, I. et al. bioRxiv Preprint at https://www.biorxiv.org/content/early/2018/07/08/364950 (2018).
Xing, E. P., Jordan, M. I. & Karp, R. M. in ICML ‘01 Proceedings of the Eighteenth International Conference on Machine Learning (eds Brodley, C. E. & Pohoreckyj Danyluk, A.) 601–608 (Morgan Kaufmann, San Francisco, 2001).
Shevade, S. K. & Keerthi, S. S. Bioinformatics 19, 2246–2253 (2003).
Trapnell, C. et al. Nat. Biotechnol. 32, 381–386 (2014).
Zheng, G. X. et al. Nat. Commun. 8, 14049 (2017).
Macosko, E. Z. et al. Cell 161, 1202–1214 (2015).
Bray, N. L., Pimentel, H., Melsted, P. & Pachter, L. Nat. Biotechnol. 34, 525–527 (2016).
Nicolae, M., Mangul, S., Măndoiu, I. I. & Zelikovsky, A. Algorithms Mol. Biol. 6, 9 (2011).
Ntranos, V., Kamath, G. M., Zhang, J. M., Pachter, L. & Tse, D. N. Genome Biol. 17, 112 (2016).
Yi, L., Pimentel, H., Bray, N. L. & Pachter, L. Genome Biol. 19, 53 (2018).
Peterson, V. M. et al. Nat. Biotechnol. 35, 936–939 (2017).
Byrne, A. et al. Nat. Commun. 8, 16027 (2017).
10x Genomics. Single cell gene expression datasets. 10x Genomics Support https://support.10xgenomics.com/single-cell-gene-expression/datasets (2018).
Wolf, F. A., Angerer, P. & Theis, F. J. Genome Biol. 19, 15 (2018).
Bradley, R. K. et al. PLoS Comput. Biol. 5, e1000392 (2009).
Petropoulos, S. et al. Cell 165, 1012–1026 (2016).
Conway, J. R., Lex, A. & Gehlenborg, N. Bioinformatics 33, 2938–2940 (2017).
Love, M. I., Huber, W. & Anders, S. Genome Biol. 15, 550 (2014).
Li, B. & Dewey, C. N. BMC Bioinformatics 12, 323 (2011).
Zappia, L., Phipson, B. & Oshlack, A. Genome Biol. 18, 174 (2017).
Satija, R., Farrell, J. A., Gennert, D., Schier, A. F. & Regev, A. Nat. Biotechnol. 33, 495–502 (2015).
Soneson, C., Love, M. I. & Robinson, M. D. F1000Res. 4, 1521 (2015).
Acknowledgements
We thank N. Bray, J. Gehring and V. Svensson for discussion and comments on the manuscript, and H. Pimentel for assisting with the simulations. We thank A. Butler and R. Satija for implementing this method in Seurat. V.N., L.Y. and L.P. are partially funded by NIH R012017-0569.
Author information
Authors and Affiliations
Contributions
V.N. developed the model during discussions with L.Y. and L.P, and analyzed the 10x PBMC dataset. L.Y. performed the simulations and analyzed the embryo SMART-Seq dataset. P.M. developed kallisto genomebam and assisted with analysis. All authors contributed extensively to the interpretation of the results and writing of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Ntranos, V., Yi, L., Melsted, P. et al. A discriminative learning approach to differential expression analysis for single-cell RNA-seq. Nat Methods 16, 163–166 (2019). https://doi.org/10.1038/s41592-018-0303-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-018-0303-9
This article is cited by
-
MENDER: fast and scalable tissue structure identification in spatial omics data
Nature Communications (2024)
-
Transient expression of the neuropeptide galanin modulates peripheral‑to‑central connectivity in the somatosensory thalamus during whisker development in mice
Nature Communications (2024)
-
Uniform quantification of single-nucleus ATAC-seq data with Paired-Insertion Counting (PIC) and a model-based insertion rate estimator
Nature Methods (2024)
-
Sox9 regulates alternative splicing and pancreatic beta cell function
Nature Communications (2024)
-
Altered microbial bile acid metabolism exacerbates T cell-driven inflammation during graft-versus-host disease
Nature Microbiology (2024)