Abstract
We report an intensiometric, near-infrared fluorescent, genetically encoded calcium ion (Ca2+) indicator (GECI) with excitation and emission maxima at 678 and 704 nm, respectively. This GECI, designated NIR-GECO1, enables imaging of Ca2+ transients in cultured mammalian cells and brain tissue with sensitivity comparable to that of currently available visible-wavelength GECIs. We demonstrate that NIR-GECO1 opens up new vistas for multicolor Ca2+ imaging in combination with other optogenetic indicators and actuators.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The NIR-GECO1 gene sequence is available through GenBank (submission no. MK134690). pDuEx2-NIR-GECO1 (plasmid no. 113,680) and pAAV-hSyn-NES-NIR-GECO1 (plasmid no. 113,683) are available via Addgene according to the terms of the Uniform Biological Material Transfer Agreement. Source data for Figs. 1–3 and Supplementary Figs. 5–14 are available online.
References
Tsien, R. Y. Annu. Rev. Biochem. 67, 509–544 (1998).
Shu, X. et al. Science 324, 804–807 (2009).
Rodriguez, E. A. et al. Nat. Methods 13, 763–769 (2016).
Piatkevich, K. D. et al. Biophys. J. 113, 2299–2309 (2017).
Shcherbakova, D. M., Stepanenko, O. V., Turoverov, K. K. & Verkhusha, V. V. Trends Biotechnol. 36, 1230–1243 (2018).
Yu, D. et al. Nat. Methods 12, 763–765 (2015).
Wagner, J. R., Zhang, J., Brunzelle, J. S., Vierstra, R. D. & Forest, K. T. J. Biol. Chem. 282, 12298–12309 (2007).
Tian, L. et al. Nat. Methods 6, 875–881 (2009).
Shcherbakova, D. M. & Verkhusha, V. V. Nat. Methods 10, 751–754 (2013).
Piatkevich, K. D. et al. Nat. Chem. Biol. 14, 352–360 (2018).
Inoue, M. et al. Nat. Methods 12, 64–70 (2015).
Shemesh, O. A. et al. Nat. Neurosci. 20, 1796–1806 (2017).
Klapoetke, N. C. et al. Nat. Methods 11, 338–346 (2014).
Wu, J. et al. ACS Chem. Neurosci. 4, 963–972 (2013).
Depry, C., Allen, M. D. & Zhang, J. Mol. Biosyst. 7, 52–58 (2011).
Harada, K. et al. Sci. Rep. 7, 7351 (2017).
Chen, T.-W. W. et al. Nature 499, 295–300 (2013).
Ohkura, M., Sasaki, T., Kobayashi, C., Ikegaya, Y. & Nakai, J. PLoS ONE 7, e39933 (2012).
Dana, H. et al. eLife 5, e12727 (2016).
Hochbaum, D. R. et al. Nat. Methods 11, 825–833 (2014).
Gambetta, G. A. & Lagarias, J. C. Proc. Natl Acad. Sci. USA 98, 10566–10571 (2001).
Wu, J. et al. Nat. Commun. 5, 5262 (2014).
Heckman, K. L. & Pease, L. R. Nat. Protoc. 2, 924–932 (2007).
Cirino, P. C., Mayer, K. M. & Umeno, D. in Directed Evolution Library Creation: Methods and Protocols (eds Arnold, F. H. & Georgiou, G.) 3–9 (Humana, Totowa, NJ, 2003).
Lin, J. Y. et al. Neuron 79, 241–253 (2013).
Pologruto, T. A., Sabatini, B. L. & Svoboda, K. Biomed. Eng. Online. 2, 13 (2003).
Krzywinski, M. & Altman, N. Nat. Methods 11, 119–120 (2014).
Acknowledgements
The authors thank the University of Alberta Molecular Biology Services Unit; Y. Li, H. Zhou and A. Aggarwal, for technical support; A. Holt for providing access to the stopped-flow spectrophotometer; and M. Vanni, T. Murphy, A. Nimmerjahn and S. Chen for preliminary AAV testing. We thank M.-E. Paquet at the University of Laval Molecular Tools Platform and the Janelia Research Campus (JRC) Virus core for AAV production, We thank V. Rancic and the JRC Histology group for preparing cultured neurons. We thank D. Park and H.J. Suk for help with characterization of NIR-GECO1 in brain-slice and two-photon imaging. We thank M. Reiss for assistance with the mouse handling, X.L. Deán-Ben for help with the in vivo mesoscale data analysis, M. Davidson and X. Shu for the mIFP gene, and E. Rodriguez for the smURFP gene. Work in R.E.C.’s lab was supported by grants from NSERC (RGPIN 288338-2010), CIHR (MOP 123514 and FS 154310), Brain Canada and NIH (UO1 NS090565). D.R. acknowledges support from the European Research Council (ERC-2015-CoG-682379). The work of D.R. and S.S was also supported by the NIH (R21-EY026382 and UF1-NS107680). Work in J.Z.’s lab was supported by NIH (R01-DK073368 and R35-CA197622). E.S.B. was supported by J. Doerr, the HHMI-Simons Faculty Scholars Program, the Open Philanthropy Project, Human Frontier Science Program (RGP0015/2016), US Army Research Laboratory and the US Army Research Office (W911NF1510548), US-Israel Binational Science Foundation (2014509) and NIH (2R01-DA029639 and 1R01-GM104948).
Author information
Authors and Affiliations
Contributions
Y.Q. developed NIR-GECO1 and performed in vitro characterization. Y.Q., K.D.P., A.S.A. and M.H.M. performed characterization in hippocampal neurons. K.D.P. and M.H.M. characterized NIR-GECO1 in intact brain slices. B.M.L. and S.G. performed in vivo mesoscale imaging. S.M. performed live-cell imaging in MIN6 β-cells. R.S.M. and M.D. measured two-photon spectra. W.Z. built the pcDuEx2 vector. Y.C. and J.W. worked on development of the smURFP-based GECI. M.D., T.E.H., J.Z., E.R.S., S.S., D.R., E.S.B. and R.E.C. supervised research. All authors were involved in data analysis. Y.Q., K.D.P. and R.E.C. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The University of Alberta has non-exclusively licensed NIR-GECO1 to LumiSTAR Biotechnology.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 The topology of mIFP (i.e., bacteriophytochrome).
Scheme is based on alignment of the sequence of mIFP (320 residues; GenBank accession number AKH03689.1; Nat. Methods 12, 763–765, 2015), with the crystal structure of the chromophore-binding domain of Deinococcus radiodurans BphP (PDB 2O9B; J. Biol. Chem. 282, 12298–12309, 2007). Representation is adapted from Takala et al. (Nature 509, 245–248; 2014), with β-strands represented as arrows and α-helical regions represented as cylinders. The PAS domain is colored in light green, and the BV-binding GAF domain is colored in light blue, as in Fig. 1a,b. The approximate position of the bound BV is represented by a magenta structure. Numbers at the ends of β-strands correspond to mIFP numbering (see Supplementary Figs. 2c, 3 and 4), based on alignment with the crystal structure. To engineer NIR-GECO1, 5 residues (171–175, DEEGN) in the loop between the first two β-strands of the GAF domain were initially replaced with a 182-residue CaM-RS20 domain (a 3-residue linker followed by 147-residue CaM followed by a 5-residue linker followed by 23-residue RS20 followed by a 4-residue linker). Systematic optimization of the insertion site to improve the Ca2+-dependent fluorescence change led to the deletion of residues 176G and 177E of mIFP, resulting in an overall replacement of 7 residues (171–177, DEEGNGE) with the CaM-RS20 domain.
Supplementary Figure 2 Directed evolution of NIR-GECO1 by library screening.
(a) Representation of the pcDuEx2 vector used for expression of genes in both bacteria and mammalian cells. Cytomegalovirus (CMV) promoter is used for mammalian expression, while the Tac promotor (a hybrid promoter derived from the trp and lac promoters) is used for bacterial expression. HO1 is expressed in bacteria but, because of the presence of a stop codon after NIR-GECO1 and the lack of a promoter before HO1, it is not expressed in mammalian cells. RBS, ribosome binding site; HO1, heme-oxygenase 1. (b) Workflow of the screening process. Briefly, E. coli DH10B was transformed with a gene library in pcDuEx2 and grown on LB plates, and then bright colonies were picked and cultured. Proteins were extracted from overnight cultures of bacteria and then tested for fluorescence and Ca2+ response in 384-well plates. Variants with reasonable brightness and Ca2+ response were selected, and the corresponding plasmids were purified. HeLa cells were transfected with the selected plasmids, and live-cell fluorescence imaging was used to evaluate both brightness and Ca2+ response. HeLa cells were not supplemented with BV. (c) Mutations of NIR-GECO1 acquired during directed evolution.
Supplementary Figure 3 Lineage of improved NIR-GECO variants.
Key mutations included deletion of 252aC and 252bE, which substantially improved the Ca2+-dependent fluorescence change; F184I and I302N, which increased affinity for Ca2+; and W350R and R357C, A455V which substantially improved the brightness. Footnotes: 1This residue was deleted in round 4. 2These mutations were reversions.
Supplementary Figure 4 Sequence alignment of NIR-GECO1 and mIFP.
Single-amino-acid changes relative to mIFP and calmodulin are highlighted with a magenta background. PAS domain, GAF domain, linkers, calmodulin, and RS20 are shown as light green, light blue, black, brown, and yellow, respectively, consistent with domain colors in Fig. 1a,b and Supplementary Fig. 1.
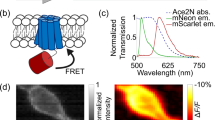
Supplementary Figure 5 Attempted engineering of a smURFP-based Ca2+ indicator.
Using methods analogous to those used to develop NIR-GECO1, we attempted to engineer a smURFP-based (Nat. Methods 13, 763–769; 2016) Ca2+ indicator. During each round of screening, the protein was tested for Ca2+-dependent fluorescent response in crude bacterial lysate prepared with B-PER protein extraction reagent (Thermo Fisher). Under these conditions, this indicator exhibited a substantial fluorescence increase upon binding Ca2+. Unfortunately we were unable to functionally express this indicator in mammalian cells. (a) Schematic representation of the smURFP-based Ca2+ indicator structure. (b) Sequence of the indicator after the eighth round of directed evolution. (c) Schematic representation of the protein structure and response. CaM-RS20 was inserted between the fourth and fifth helix of smURFP. The best variant exhibited an approximately eightfold change in fluorescence intensity (Ca2+-bound/Ca2+-free) at the maximum emission (670 nm). (d) Emission spectra for the protein after the eighth round, normalized to the Ca2+-free state.
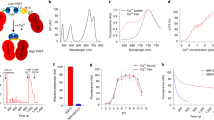
Supplementary Figure 6 Additional in vitro characterization of NIR-GECO1.
(a) pH titration curves of NIR-GECO1 in the presence and absence of Ca2+. n = 3 independent experiments; values are mean ± s.d. (b) Ca2+ dissociation kinetics of NIR-GECO1 (magenta) and GCaMP6s (green).
Supplementary Figure 7 Increasing intracellular BV concentration has a modest effect on NIR-GECO1 brightness.
(a) Representative fluorescence traces of NIR-GECO1 (top) and NIR-GECO1 supplied with 25 μM exogenous BV (bottom) in response to neuronal spontaneous activities. n = 51 neurons for NIR-GECO and n = 39 neurons for NIR-GECO1 + BV. (b) Quantification of –ΔF/F0 corresponding to the experiment of a. Values are 16 ± 6% for NIR-GECO1 and 20 ± 8% for NIR-GECO1 + BV (mean ± s.d.). (c) Representative wide-field fluorescence images of neurons expressing NIR-GECO1 (left) and NIR-GECO1-T2A-HO1 (right). The human HO1 gene was used. Scale bar, 20 μm. (d) Relative normalized fluorescence of NIR-GECO1 (n = 15 neurons) and NIR-GECO1-T2A-HO1 (n = 15 neurons). Values are 18.3 ± 10.2 (a.u.) for NIR-GECO1 and 27.1 ± 7.0 (a.u.) for NIR-GECO1-T2A-HO1 (mean ± s.d.). Fluorescence was normalized by coexpression of EGFP (NIR channel, 650/60 nm Ex and 720/60 nm Em; green channel, 490/15 nm Ex and 525/50 nm Em). Box plots are used where the top and bottom horizontal lines mark the 25th and 75th percentiles for the data; whiskers extend to the maximum and minimum for the data; and the black horizontal bar is the median.
Supplementary Figure 8 Characterization of NIR-GECO1 in cultured neurons and in intact brain tissues.
(a) Representative traces of single-trial NIR-GECO1 fluorescence responses to field stimulation in cultured rat hippocampal neurons. (b) Representative confocal images of neurons in L2/3 of motor cortex expressing NIR-GECO1 (641 nm Ex and 664LP Em; n = 4 slices from 2 mice). Such neurons were imaged during electrophysiological current injections as in c. (c) Representative single-trial wide-field optical recording of NIR-GECO1 fluorescence responses (magenta; 631/28 nm Ex and 664LP Em; acquisition rate 50 Hz) to 1, 5, 10, and 20 action potentials trains evoked by current injections in neurons in L2/3 of motor cortex (as in b; n = 6 neurons from 4 mice at P11–22). Patch voltage is shown in black.
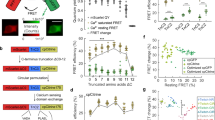
Supplementary Figure 9 Comparison of NIR-GECO1, NES-NIR-GECO1 and GCaMP6s.
(a) Response amplitude of NIR-GECO1 (n = 55 neurons) and NES-NIR-GECO1 (n = 147 neurons) as a fraction of GCaMP6s (n = 31 neurons). The average –ΔF/Fmin of NIR-GECO1 was 2.4 ± 0.12%, 3.4 ± 0.16%, 6.6 ± 0.29%, 11 ± 0.44%, 17 ± 0.65%, 27 ± 0.9%, 43 ± 1.4%, 60 ± 2.0%, 77 ± 3.0%, and 94 ± 4.2% for 1, 2, 3, 5, 10, 20, 40, 80, 120 and 160 APs, respectively. Relative to GCaMP6s, the –ΔF/Fmin of NIR-GECO1 was 10% of GCaMP6s for 1 and 2 APs and increased to 15% and 13% for 3 and 5 APs and then went down to 7% for APs from 10 to 160 APs. Elsewhere in the paper we have consistently used ΔF/F0 to describe fluorescence changes. Here we use ΔF/Fmin to enable the values for NIR-GECO1 and GCaMP6s to be easily compared. (b) Signal-to-noise ratio (SNR) of NIR-GECO1 and NES-NIR-GECO1 compared to GCaMP6s. The SNR of NIR-GECO1 was 26.7 ± 1.98, 34.4 ± 2.15, 62.2 ± 3.42, 98.6 ± 6.37, 145 ± 9.29, 185 ± 10.9, 256 ± 15.3, 302 ± 18.0, 311 ± 16.9 and 335 ± 18.8 for 1, 2, 3, 5, 10, 20, 40, 80, 120 and 160 APs, respectively. Relative to GCaMP6s, the SNR of NIR-GECO1 was 20% of GCaMP6s for APs from 1 to 5 and then goes down to 12% to 4% of GCaMP6s for APs from 10 to 100. (c) Fluorescence rise time of NIR-GECO1 and NES-NIR-GECO1 (actually a fluorescence decrease) compared to GCaMP6s for binding of Ca2+. The average rise time of NIR-GECO1 was 0.94 ± 0.033 s, 1.4 ± 0.038 s, 1.4 ± 0.044 s for 1, 10 and 40 APs. (d) Fluorescence decay time of NIR-GECO1 and NES-NIR-GECO1 (actually a fluorescence increase) compared to GCaMP6s for dissociation of Ca2+. The average decay time of NIR-GECO1 was 2.8 ± 0.21 s, 3.9 ± 0.12 s, 4.6 ± 0.11 s for 1, 10 and 40 APs. For a–d, the NIR-GECO1 data are identical to data represented in Fig. 1i–l. For a–d, center values are the mean, and error bars are s.e.m. n = 55 neurons for NIR-GECO1 and n = 31 neurons for GCaMP6s. (e) Representative wide-field fluorescence images of coexpressed NIR-GECO1 (left) and GCaMP6s (right). n = 5 neurons from two cultures. Scale bar, 20 μm. (f) Spontaneous Ca2+ oscillations in dissociated cortical neurons coexpressing NIR-GECO1 and GCaMP6s (NIR channel, 650/60 nm Ex and 720/60 nm Em; green channel, 490/15 nm Ex and 525/50 nm Em; acquisition rate is 1 Hz). Also shown is ΔR/R0, where R is the normalized GCaMP6s intensity divided by normalized NIR-GECO1 intensity. n = 5 neurons from two cultures.
Supplementary Figure 10 In vivo mesoscale imaging of footshock responses in mice using NIR-GECO1.
(a) Mesoscale fluorescence images (671 nm Ex and 721/42 nm Em) of the mouse sensorimotor cortex injected with AAV2/9-hSyn1-NIR-GECO1 (as in Fig. 2e–g). Left, negative control with no viral expression on the right side of mouse brain (PBS injection). Middle, viral expression of NIR-GECO1 on the right side of mouse brain. Right, viral expression of NIR-GECO1 on the left side of mouse brain. Scale bar, 2 mm. (b) Fluorescence response of NIR-GECO1 in response to a paw stimulation paradigm with a single 50-ms pulse (0.5 mA). As in Fig. 2e, each gray line represents the averaged response of a mouse across 19 cycles, and the blue line represents the mean response from all 3 mice (n = 3, or 3 × 19 = 57 cycles). (c) Activation map of mouse brain before stimulation. The estimated brain outline was manually superimposed onto the fluorescence images to facilitate determining the site of injection and activation in relation to bregma and the sensorimotor cortex. (d) Activation map of mouse brain at max activation. Scale bar, 2 mm. OB, olfactory bulb; CB, cerebellum; L/RC, left or right cortex.
Supplementary Figure 11 Control experiments for in vivo mesoscale imaging using GCaMP6s and miRFP.
(a) Comparative fluorescence images of a transgenic Bl6-GCaMP6s mouse and FoxN1 nude NIR-GECO1, miRFP and PBS-injected mice. All fluorescence images are normalized to the GCaMP image shown. Direct comparison of the NIR-GECO1 and miRFP brightness is complicated by the fact that the AAVs were different serotypes (AAV2/9 and AAV2, respectively) and the NIR-GECO1 stock had 10 × more genome copies/mL. Also shown is a representative white light image of the imaging area. Scale bar, 2 mm. (b, c) Positive control experiment with imaging of GCaMP6s in response to paw stimulations. For b, a stimulation paradigm of a single 50-ms pulse (0.5 mA) was used. For c, a stimulation paradigm of 10 pulses in 700 ms (0.5 mA, 20 ms on and 50 ms off) was used. The mean value from 3 mice is shown in each case. NIR-GECO1 curves are the same as in Supplementary Fig. 10b and Fig. 2e. (d, e) A negative control experiment with imaging of miRFP fluorescence in response to paw stimulations. Stimulations in d and e are the same as in b and c, respectively. Orange line represents the mean value from 3 mice, and gray lines represent the average response of 1 mouse across 19 cycles. (f, g) A negative control with fluorescence imaging PBS-injected mouse in response to paw stimulations. Filter set is the same as for NIR-GECO1. The green line represents the mean value of 3 measurements (an average of 19 cycles) from 2 mice, and the gray lines represent the average response averaged for 1 mouse measured once and 1 mouse measured twice. Stimulations in f and g are the same as in b and c, respectively. The GCaMP6s mice are a transgenic line, and the mice were approximately 2 months older than other mice used in these experiments.
Supplementary Figure 12 Two-photon fluorescence microscopy of NIR-GECO1.
BV-FPs can be visualized using two-photon fluorescence microscopy (Biophys. J. 113, 2299–2309; 2017), a widely used technique for in vivo Ca2+ imaging. (a) Relative normalized fluorescence and (b) raw photobleaching curves for iRFP682 (blue), miRFP (cyan), and NIR-GECO1 (magenta) in cultured mouse neurons (n = 184, 106, and 77 cells, respectively, from one culture) under 880-nm two-photon excitation and 4.05 mW of total light power. For a, a box plot with notches is used as described in Fig. 1g. (c) Representative fluorescence recording of 4-aminopyridine (1 mM final concentration)-evoked neuronal activity using NIR-GECO1 and GCaMP6f fluorescence under 880-nm two-photon excitation. Excitation for both NIR-GECO1 and GCaMP6f was 880 nm, and emission filters for NIR-GECO1 and GCaMP6s were 705/90 nm and 518/45 nm, respectively. (d) Representative two-photon fluorescence images of cultured mouse neurons expressing iRFP682 (left), miRFP (middle) and NIR-GECO1 (right), under 880-nm two-photon excitation (731/137 nm Em; n = 184, 106, and 77 cells, respectively, from one culture). Scale bar, 50 μm. (e) Two-photon fluorescence images of live mouse brain slice coexpressing NIR-GECO1 (left; magenta) and GCaMP6s (middle; green) under 880-nm excitation (right, overlay). n = 2 slices from one mouse. Scale bar, 50 μm. (f) In vivo two-photon microscopy of NIR-GECO1-expressing neurons in mouse primary visual cortex. n = 3 fields of view from one mouse. Scale bar, 50 μm.
Supplementary Figure 13 Combined use of channelrhodopsins (CoChR and CheRiff) and NIR-GECO1 for optogenetic stimulation and imaging of activity.
Representative wide-field fluorescent images of cultured hippocampal mouse neurons expressing NIR-GECO1 (top; magenta; 631/28 nm Ex and 664LP Em), and both NIR-GECO1 (magenta; 631/28 nm Ex and 664LP Em) and CoChR-EGFP (Nat. Methods 11, 338–346, 2014; Nat. Neurosci. 20, 1796–1806, 2017) (green; bottom; 475/34 nm Ex and 525/50 nm Em). Scale bar, 50 µm. (b) Representative single-trial traces for single neurons (as in a; n = 45 and 93 neurons for NIR-GECO1 and NIR-GECO1 + CoChR-EGFP, respectively, from two cultures) illuminated with 562/40 nm (orange arrows) or 475/34 nm (blue arrows) at indicated light intensities with 200-ms duration per pulse. (c) Quantification of NIR-GECO1 fluorescence changes in response to activation of CoChR under the conditions described in b (n = 45 and 93 neurons for NIR-GECO1 and NIR-GECO1 + CoChR-EGFP, respectively, from 2 cultures). Box plots with notches are used as described in Fig. 1g. (d) Image of cultured rat hippocampal neurons expressing CheRiff-EGFP (Nat. Methods 11, 825–833; 2014) (green, 480 nm Ex and 525/36 nm Em) and NIR-GECO1 (magenta, 640 nm Ex and 705/50 nm Em). CheRiff is localized to the plasma membrane, whereas NIR-GECO1 fills the cytoplasm and nucleus of the neurons. Scale bar, 20 µm. (e) NIR-GECO1 fluorescence traces from 4 neurons, indicated with correspondingly colored arrows in d, in response to optical stimulation. Blue bar indicates a 10-ms blue light (490/20 nm at 4 mW/mm2) illumination.
Supplementary Figure 14 Additional representative single-cell traces for multiplexed imaging of MIN6 β-cells.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–14, Supplementary Tables 1 and Supplementary Notes 1–5
Supplementary Video 1
In vivo mesoscale imaging of footshock responses in mouse sensorimotor cortex: as in Fig. 2e–g.
Supplementary Video 2
In vivo mesoscale imaging of footshock responses in mice using NIR-GECO1: as in Supplementary Fig. 10b–d.
Supplementary Video 3
Multicolor imaging of cell signaling with NIR-GECO1. Representative videos, simultaneously recorded in different color channels, of a MIN6 β cell coexpressing Pink Flamindo (fluorescence intensity), NIR-GECO1 (fluorescence intensity), and AKAR4 (pseudocolored FRET emission ratio). Traces for this cell are provided in Supplementary Fig. 14d. Traces for additional cells are provided in Fig. 3g and Supplementary Fig. 14a–c.
Supplementary Video 4
Three-color fluorescence imaging of spontaneous neuronal activity in dissociated neurons using NIR-GECO1, GCaMP6f, and RCaMP1.07: as in Fig. 3h.
Source Data
Rights and permissions
About this article
Cite this article
Qian, Y., Piatkevich, K.D., Mc Larney, B. et al. A genetically encoded near-infrared fluorescent calcium ion indicator. Nat Methods 16, 171–174 (2019). https://doi.org/10.1038/s41592-018-0294-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-018-0294-6
This article is cited by
-
PMF-seq: a highly scalable screening strategy for linking genetics to mitochondrial bioenergetics
Nature Metabolism (2024)
-
Widefield imaging of rapid pan-cortical voltage dynamics with an indicator evolved for one-photon microscopy
Nature Communications (2023)
-
Quantitative assessment of near-infrared fluorescent proteins
Nature Methods (2023)
-
Chemigenetic indicators based on synthetic chelators and green fluorescent protein
Nature Chemical Biology (2023)
-
Structural and functional imaging of brains
Science China Chemistry (2023)