Abstract
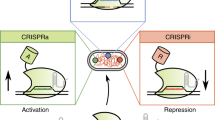
CRISPR–Cas9-based combinatorial perturbation approaches for orthogonal knockout and gene activation have been impeded by complex vector designs and co-delivery of multiple constructs. Here, we demonstrate that catalytically active CRISPR–Cas12a fused to a transcriptional-activator domain enables flexible switching between genome editing and transcriptional activation by altering guide length. By leveraging Cas12a-mediated CRISPR-RNA array processing, we illustrate that Cas12a-VPR enables simplified multiplexed knockout and transcriptional activation in vitro and in vivo.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Costanzo, M., Baryshnikova, A., Myers, C. L., Andrews, B. & Boone, C. Curr. Opin. Biotechnol. 22, 66–74 (2011).
Zalatan, J. G. et al. Cell 160, 339–350 (2015).
Gao, Y. et al. Nat. Methods 13, 1043–1049 (2016).
Du, D. et al. Nat. Methods 14, 577–580 (2017).
Han, K. et al. Nat. Biotechnol. 35, 463–474 (2017).
Najm, F. J. et al. Nat. Biotechnol. 36, 179–189 (2018).
Boettcher, M. et al. Nat. Biotechnol. 36, 170–178 (2018).
Kiani, S. et al. Nat. Methods 12, 1051–1054 (2015).
Dahlman, J. E. et al. Nat. Biotechnol. 33, 1159–1161 (2015).
Zetsche, B. et al. Nat. Biotechnol. 35, 31–34 (2017).
Zhong, G., Wang, H., Li, Y., Tran, M. H. & Farzan, M. Nat. Chem. Biol. 13, 839–841 (2017).
Zetsche, B. et al. Cell 163, 759–771 (2015).
Kleinstiver, B. P. et al. Nat. Biotechnol. 34, 869–874 (2016).
Kim, H. K. et al. Nat. Methods 14, 153–159 (2017).
Singh, D. et al. Proc. Natl. Acad. Sci. USA 115, 5444–5449 (2018).
Tak, Y. E. et al. Nat. Methods 14, 1163–1166 (2017).
Tschaharganeh, D. F. et al. Cell 158, 579–592 (2014).
Perez-Pinera, P. et al. Nat. Methods 10, 973–976 (2013).
Kim, H. K. et al. Nat. Biotechnol. 36, 239–241 (2018).
Labun, K., Montague, T. G., Gagnon, J. A., Thyme, S. B. & Valen, E. Nucleic Acids Res. 44, W272–W276 (2016).
Acknowledgements
We thank all members of the laboratory of D.F.T. for insightful comments on this work. We thank the DKFZ Genomics Core Facility, the DKFZ Central Animal Laboratory, and the CMCP of the Institute of Pathology Heidelberg for excellent technical support. This work was supported by the Helmholtz Association (VH-NG-1114), the Helmholtz Association ERC Recognition Award, and the German Research Foundation (DFG) project B05, SFB/TR 209 ‘Liver Cancer’ (all awarded to D.F.T.). We thank F. J. Sánchez-Rivera (Cancer Biology and Genetics, Memorial Sloan Kettering Cancer Center), G. Church (Department of Genetics, Harvard Medical School), E. Welker (Institute of Biochemistry, Biological Research Centre, Hungarian Academy of Sciences), F. Zhang (Department of Biological Engineering, Massachusetts Institute of Technology), and D. Trono (School of Life Sciences, Ecole Polytechnique Federale de Lausanne (EPFL)) for providing reagents.
Author information
Authors and Affiliations
Contributions
M.B. performed experiments, analyzed data, designed the study, and wrote the manuscript. S.R., A.Y.S., A.M.H., A.C.G., L.S., and L.W. performed experiments. D.F.T. performed experiments, analyzed data, designed the study, and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Orthogonal gene control with catalytically active Cas12a-VPR.
(a) Schematic depicting the strategy for Cas12a-VPR in which 20-nt guides produce indels and 15-nt guides allow transcriptional activation. (b) Overview of vector constructs. U6, Pol III promoter; NLS, nuclear localization signal;DR, direct repeat sequence; LTR, long terminal repeat; WPRE, woodchuck hepatitis virus post-transcriptional regulatory element;IR, inverted repeat;pA, polyA; PGK, phosphoglycerate kinase 1 promoter; puro = puromycin resistance; Blas = blasticidine resistance; EFs, truncated human elongation factor-1; EF1, human elongation factor-1 promoter; Mir30-shRNA-REN, microRNA-adapted short hairpin RNA targeting renilla; GFP, green fluorescent protein; BFP, blue fluorescent protein; VPR, herpes simplex virus-derived VP16 activator, human NF-KB p65 activation domain, Epstein–Barr-virus-derived R transactivator (Rta)).
Supplementary Figure 2 AsCas12a-VPR-mediated reporter activation is dictated by guide length.
(a) Schematic of the TRE-RFP reporter system. Mouse cells stably expressing TRE-driven RFP and the reverse Tetracycline transactivator (rtTA3) (Tet-on system) are used. Each TRE-guide has seven potential target sites within the TRE promoter. (b) Schematic of the reporter cell line employed. A doxycycline-inducible TRE-RFP-shp53 reporter cell line was used. Dox withdrawal results in growth arrest and loss of RFP expression. AsCas12a-VPR-mediated transcriptional activation using TRE-targeting guides induces RFP expression and rescues cell gowth in off Dox conditions. Dox = doxycycline. (c) PCR of the TRE promoter using genomic DNA obtained 10 days post-lentiviral transduction. A doxycycline-inducible TRE-RFP reporter was used to assess transcriptional activation in mouse cells stably expressing AsCas12a-VPR (right) or AsCas12a (left) and crRNAs. TRE-guide length was varied from 20-nt to 12-nt. crRNAs were lentivirally-transduced. ctrl = Rosa26-guide. N = 2 independent experiments; Representative images are shown. (d) Flow cytometry-based RFP quantification and in reporter cell lines stably expressing AsCas12a-VPR (left) or AsCas12a (right) transduced with crRNAs expressing different guide lengths (20-nt to 12-nt) targeting the TRE promoter in off Dox and on Dox conditions. Mean ± s.d.; N = 3 independent experiments. (e) Cell growth as determined by colony formation using crystal violet (CV). Reporter cell lines stably expressing AsCas12a-VPR (left) or AsCas12a (right) were transduced with crRNAs expressing different guide lengths (20-nt to 12-nt) targeting the TRE promoter and grown under off DOX conditions for 14d. ctrl = Rosa26-guide. N = 2 independent experiments. Representative images are shown.
Supplementary Figure 3 Comparison of transcriptional-activation capabilities of AsCas12a-VPR with LbCas12a-VPR and deadLbCas12a-VPR.
a) Flow cytometry-based RFP quantification in murine reporter cell lines stably expressing either AsCas12a-VPR1 or LbCas12a-VPR and crRNAs harboring either the As-DR or Lb-DR. TRE-guide length was varied from 20-nt to 12-nt. ctrl = Rosa26-guide. Cells were cultivated off Dox. Mean ± s.d.; N = 3 independent experiments. b) Flow cytometry-based RFP quantification in murine reporter cell lines stably expressing deadLbCas12a-VPR. TRE-guide length was varied from 20-nt to 12-nt. crRNAs harboring the Lb-DR were used. ctrl = Rosa26-guide. Cells were cultivated off Dox. Mean ± s.d.; N = 3 independent experiments.
Supplementary Figure 4 Comparison of transcriptional-activation capabilities of AsCas12a-VPR with other CRISPR effectors.
Flow cytometry-based RFP quantification in reporter cell lines stably expressing indicated CRISPR-effectors. A Doxycycline-inducible TRE-RFP-shp53 reporter was used to assess transcriptional activation. Mouse cells stably expressing indicated CRISPR-effectors were transduced with 15-nt or 20-nt TRE-guides to activate RFP. Cas12a and Cas9 guides were position matched and embedded in respective expression vectors. Mean ± s.d.; N = 3 independent experiments.
Supplementary Figure 5 Transcriptional activation of endogenous genes with catalytically active AsCas12a-VPR and 15-nt guides, and comparison to LbCas12a-VPR or deadLbCas12a-VPR.
a) Relative mRNA expression after co-transfection of HEK293T cells with AsCas12a-VPR and 15-nt guides targeting the promoter region of indicated genes. N = 2 independent experiments. b) Relative mRNA expression after co-transfection of HEK293T cells with either AsCas12a-VPR, LbCas12a-VPR or deadLbCas12a-VPR and crRNAs. crRNAs were targeted to the promoter of TTN (left) or NPY1R (right). For TTN, N = 3 independent experiments. For NPY1R, N = 2 independent experiments.
Supplementary Figure 6 Exon-targeting 20-nt guides do not induce levels of transcriptional activation comparable to those of 15-nt and 20-nt guides targeted to promoter regions.
(a) Schematic of the approach to investigate the impact of 20-nt exon targeting guides on transcriptional activation. A representative gene model is shown; not drawn to scale. 15-nt as well as 20-nt guides targeting the promoter region or 20-nt guides targeting different genomic loci within exons were used. Three different qPCR primer pairs were used located either before or after the respective genomic loci of exon-targeting guides in order to measure transcriptional activation. (b) Relative mRNA expression after co-transfection of HEK293T cells with AsCas12a-VPR and crRNAs. 15-nt as well as 20-nt guides targeting the promoter region or 20-nt guides targeting different genomic loci within exons of TTN, NPY1R or MYOD1 were used. N = 2 independent experiments.
Supplementary Figure 7 AsCas12a-VPR and 15-nt guides induce transcriptional activation in vivo.
(a) HDTV of AsCas12a-VPR, crRNA expressing vectors, and SB into TRE-GFP reporter knock-in mice. crRNAs with a 20-nt Rosa26 ctrl-guide, as well as a 20-nt TRE-guide and the corresponding 15-nt TRE-guide were tested. 2 weeks post-HDTV, expression of GFP in livers was investigated by immunofluorescence. BFP staining indicates crRNA expression. DAPI staining visualizes nuclei. N = 3 mice per group. Representative images are shown. Scale bar = 100 µm. (b) HDTV of AsCas12a-VPR, 15-nt Krt19-guide expressing vectors, and SB into mice. Immunohistochemistry of Krt19. crRNAs with a 15-nt Rosa26 ctrl-guide, as well as 15-nt KRT19-guides were tested. Livers were analyzed 2 weeks post-HDTV. * = Krt19 positive bile duct cells/cholangiocytes; arrows = Krt19 positive hepatocytes. N = 5 animals per group. Representative results are shown. Scale bar = 20 µm.
Supplementary Figure 8 AsCas12a-VPR and 20-nt guides produce knockout phenotypes in vivo.
(a) Schematic of the reporter cell line employed for in vitro validation of Trp53-guide activity. A doxycycline-inducible TRE-RFP-shp53 reporter cell line was used. Dox withdrawal results in growth arrest. Trp53 loss-of-function rescues cell growth. (b) Cell growth as determined by colony formation using crystal violet (CV). 20-nt Trp53-guides were lentivirally delivered to mouse cells stably expressing AsCas12a-VPR. N = 2 independent experiments; Representative results are shown. (c) Relative mRNA expression after lentiviral delivery of 20-nt Trp53-guides to mouse cells stably expressing AsCas12a-VPR. Schematic of the approach showing a gene model not drawn to scale. Three different exon-targeting 20-nt Trp53-guides were used. Three different qPCR primer pairs located at varying loci were used to determine mRNA abundance. N = 2 independent experiments. (d) Cell growth as determined by colony formation using crystal violet (CV). Trp53-guide 3 of different lengths were lentivirally delivered to mouse cells stably expressing AsCas12a-VPR. N = 2 independent experiments; Representative results are shown. (e) Schematic depicting the in vivo experiment in which Trp53 loss-of-function results in liver tumor formation. HDTV of AsCas12a-VPR, 20-nt Trp53-guide crRNA expressing vector, c-Myc, and SB into mice. (f) Tumor-free survival. N = 3 animals per group. Rosa26-guide serves as control. (g) Livers taken 7–8 weeks post-HDTV. N = 3 mice per group. Representative images are shown. Scale bar = 1 cm. (h) Indel formation in the Trp53 locus as evidenced by T7EI assay. T7EI assay was performed with three tumor derived cell lines. Trp53 wt murine tumor cell lines were used as control (ctrl cells 1 and 2, respectively). N = 2 independent experiments; Representative results are shown. (i) Trp53 western blot. Cell lines derived from mouse tumors were treated with Doxorubicin to induce Trp53 expression. Trp53 protein levels were determined by Western blot. A Trp53-proficient mouse tumor cell line was used as control. N = 2 independent experiments; Representative results are shown.
Supplementary Figure 9 crRNA array-based transcriptional activation or knockout with AsCas12a-VPR.
(a) Schematic of the reporter cell line employed. A doxycycline-inducible TRE-RFP-shp53 reporter cell line was used. Dox withdrawal results in growth arrest and loss of RFP expression. Expression of a 15-nt TRE-guide induces RFP activation and rescues cell growth. Trp53 loss-of-function rescues cell growth. (b) Flow cytometry-based RFP quantification in mouse reporter cell line stably expressing AsCas12a-VPR transduced with different crRNA arrays harboring the 15-nt TRE-guide as indicated. Cells were cultivated in off Dox conditions. Mean ± s.d.; N = 3 independent experiments. (c) Cell growth as determined by colony formation using CV staining. N = 3 independent experiments; Representative results are shown. (d) Cell growth as determined by colony formation using crystal violet (CV). A 20-nt Trp53-guide was embedded in different crRNA arrays as indicated and lentivirally delivered to mouse cells stably expressing AsCas12a-VPR. N = 2 independent experiments; Representative results are shown. (e) Indel formation as detected by the T7EI assay. A 20-nt Trp53-guide was embedded in different crRNA arrays as indicated and lentivirally delivered to mouse cells stably expressing AsCas12a-VPR. N = 2 independent experiments; Representative results are shown.
Supplementary Figure 10 Simultaneous activation of endogenous genes by catalytically active AsCas12a-VPR and crRNA arrays.
Relative mRNA expression after co-transfection of HEK293T cells with AsCas12a-VPR and crRNA arrays. Multiplex expression of 15-nt guides expressed from a single array versus respective single crRNAs targeting the promoter region of MIAT, or MYOD1. Mean ± s.d.; N = 3 independent experiments.
Supplementary Figure 11 Array-based multiplexing enhances AsCas12a-VPR-mediated transcriptional activation.
Relative mRNA expression after co-transfection of HEK293T cells with AsCas12a-VPR and crRNA arrays. Multiplex expression of three 15-nt guides expressed from a single array versus respective single crRNAs targeting the promoter of TTN, or NPY1R respectively. Mean ± s.d.; N = 3 independent experiments.
Supplementary Figure 12 Orthogonal gene control of endogenous genes by catalytically active AsCas12a-VPR and crRNA arrays.
(a) Relative mRNA expression after co-transfection of HEK293T cells with AsCas12a-VPR and crRNA arrays. Multiplex expression of a 20-nt guide targeting MIAT and a 15-nt guide targeting the MYOD1 promoter (left) or a 20-nt guide targeting MYOD1 and a 15-nt guide targeting the MIAT promoter (right), using a single array versus respective single crRNAs. Mean ± s.d.; N = 3 independent experiments. (b) Indel formation after co-transfection of HEK293T cells with AsCas12a-VPR and crRNA arrays as measured by T7EI assay. Multiplex expression of a 20-nt guide targeting MIAT and a 15-nt guide targeting the MYOD1 promoter (left) or a 20-nt guide targeting MYOD1 and a 15-nt guide targeting the MIAT promoter (right), using a single array versus respective single crRNAs. N = 2 independent experiments; Representative results are shown.
Supplementary Figure 13 AsCas12a-VPR-mediated orthogonal gene control is achieved within single cells.
(a) Flow cytometry-based RFP and BFP quantification in murine reporter cell lines stably expressing AsCas12a-VPR transduced with crRNA arrays. Multiplex expression of a 20-nt guide targeting BFP and a 15-nt TRE-guide using a single array versus single crRNAs. N = 2 independent experiments; Representative results are shown. (b) Flow cytometry-based RFP and BFP quantification in murine reporter cell lines stably expressing AsCas12a-VPR transduced with crRNA arrays. Gating of relevant cell populations is depicted. N = 2 independent experiments; Representative results are shown.
Supplementary Figure 14 Simultaneous activation and orthogonal gene control of endogenous genes with catalytically active AsCas12a-VPR and lentiviral delivery of crRNAs and arrays.
(a) Estimation of lentiviral multiplicity of infection (MOI) using flow cytometry-based GFP quantification. HEK293T cells were transduced with different volumes of lentiviral supernatant and GFP expression -as a surrogate marker for MOI- was measured three days after lentiviral addition. Ctrl = HEK293T cells without addition of virus. N = 2 independent lentiviral transductions. Boxes represent the 25th, 50th and 75th percentiles; whiskers show maxima. For group “Low MOI” and “high MOI”, individual data points represent different crRNA vectors (n = 10). (b) Relative mRNA expression after lentiviral transduction of HEK293T cells with crRNA arrays using low or high MOI. HEK293T cells expressing AsCas12a-VPR were lentivirally transduced with indicated crRNAs and crRNA arrays including 15-, and 20-nt guides targeting the promoter region of TTN or NPY1R. N = 2 independent experiments. (c) Indel formation after lentiviral delivery of crRNAs and arrays to HEK293T cells expressing AsCas12a-VPR as measured by T7EI assay. Multiplex expression of a 20-nt guide targeting NPY1R and a 15-nt guide targeting the TTN promoter using a single array versus respective single crRNAs. N = 2 independent experiments. Representative results are shown.
Supplementary Figure 15 In vivo multiplexed transcriptional activation and orthogonal gene control by catalytically active AsCas12a-VPR and crRNA arrays.
HDTV of AsCas12a-VPR, crRNA array expressing vectors, and SB into mice stably expressing a TRE-GFP reporter. Arrays for multiplex expression of a 20-nt Trp53-guide combined with 15-nt Rosa26 ctrl-guide were used. GFP expression in livers taken 8 weeks post-HDTV was visualized by immunofluorescence. BFP staining indicates crRNA expression. DAPI staining visualizes nuclei. N = 5 mice per group. Representative images are shown. Scale bar = 80 µm. Indel occurrence at the Trp53 locus. N = 2 independent liver samples. Representative results are shown.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–15 and Supplementary Tables 1–5
Rights and permissions
About this article
Cite this article
Breinig, M., Schweitzer, A.Y., Herianto, A.M. et al. Multiplexed orthogonal genome editing and transcriptional activation by Cas12a . Nat Methods 16, 51–54 (2019). https://doi.org/10.1038/s41592-018-0262-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-018-0262-1
This article is cited by
-
Prime editing using CRISPR-Cas12a and circular RNAs in human cells
Nature Biotechnology (2024)
-
CRISPR-Combo–mediated orthogonal genome editing and transcriptional activation for plant breeding
Nature Protocols (2023)
-
An update on CRISPR-Cas12 as a versatile tool in genome editing
Molecular Biology Reports (2023)
-
Engineered Cas12a-Plus nuclease enables gene editing with enhanced activity and specificity
BMC Biology (2022)
-
Multiplexed genome regulation in vivo with hyper-efficient Cas12a
Nature Cell Biology (2022)