Abstract
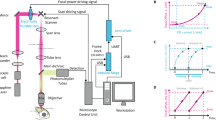
Whole-brain imaging allows for comprehensive functional mapping of distributed neural pathways, but neuronal perturbation experiments are usually limited to targeting predefined regions or genetically identifiable cell types. To complement whole-brain measures of activity with brain-wide manipulations for testing causal interactions, we introduce a system that uses measured activity patterns to guide optical perturbations of any subset of neurons in the same fictively behaving larval zebrafish. First, a light-sheet microscope collects whole-brain data that are rapidly analyzed by a distributed computing system to generate functional brain maps. On the basis of these maps, the experimenter can then optically ablate neurons and image activity changes across the brain. We applied this method to characterize contributions of behaviorally tuned populations to the optomotor response. We extended the system to optogenetically stimulate arbitrary subsets of neurons during whole-brain imaging. These open-source methods enable delineating the contributions of neurons to brain-wide circuit dynamics and behavior in individual animals.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Raw data for Fig. 1c are available at https://github.com/optofish-paper/ (downsampled in space to avoid excessive data size). Example processing steps for Python notebooks are available at the same address. Processed data for Fig. 2 are available at the same address in the form of full brain volumes.
References
Vogel, A. & Venugopalan, V. Mechanisms of pulsed laser ablation of biological tissues. Chem. Rev. 103, 577–644 (2003).
Deisseroth, K. Optogenetics: 10 years of microbial opsins in neuroscience. Nat. Neurosci. 18, 1213–1225 (2015).
Yang, W., Carrillo-Reid, L., Bando, Y., Peterka, D. S. & Yuste, R. Simultaneous two-photon imaging and two-photon optogenetics of cortical circuits in three dimensions. eLife 7, e32671 (2018).
Liu, X. et al. Optogenetic stimulation of a hippocampal engram activates fear memory recall. Nature 484, 381–385 (2012).
Chen, S., Chiu, C. N., McArthur, K. L., Fetcho, J. R. & Prober, D. A. TRP channel mediated neuronal activation and ablation in freely behaving zebrafish. Nat. Methods 13, 147–150 (2016).
Lee, J. H. et al. Global and local fMRI signals driven by neurons defined optogenetically by type and wiring. Nature 465, 788–792 (2010).
Boyden, E. S., Zhang, F., Bamberg, E., Nagel, G. & Deisseroth, K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 8, 1263–1268 (2005).
Rickgauer, J. P., Deisseroth, K. & Tank, D. W. Simultaneous cellular-resolution optical perturbation and imaging of place cell firing fields. Nat. Neurosci. 17, 1816–1824 (2014).
Packer, A. M., Russell, L. E., Dalgleish, H. W. P. & Häusser, M. Simultaneous all-optical manipulation and recording of neural circuit activity with cellular resolution in vivo. Nat. Methods 12, 140–146 (2015).
Dal Maschio, M., Donovan, J. C., Helmbrecht, T. O. & Baier, H. Linking neurons to network function and behavior by two-photon holographic optogenetics and volumetric imaging. Neuron 94, 774–789.e5 (2017).
Carrillo-Reid, L., Han, S., Yang, W., Akrouh, A. & Yuste, R. Triggering visually-guided behavior by holographic activation of 1 pattern completion neurons in cortical ensembles 2 3. Preprint at https://www.biorxiv.org/content/early/2018/08/17/394999 (2018).
Chen, X. et al. Brain-wide organization of neuronal activity and convergent sensorimotor transformations in larval zebrafish. Neuron https://doi.org/10.1016/j.neuron.2018.09.042 (2018).
Portugues, R., Feierstein, C. E., Engert, F. & Orger, M. B. Whole-brain activity maps reveal stereotyped, distributed networks for visuomotor behavior. Neuron 81, 1328–1343 (2014).
Naumann, E. A. et al. From whole-brain data to functional circuit models: the zebrafish optomotor response. Cell 167, 947–960.e20 (2016).
Grosenick, L., Marshel, J. H. & Deisseroth, K. Closed-loop and activity-guided optogenetic control. Neuron 86, 106–139 (2015).
Otchy, T. M. et al. Acute off-target effects of neural circuit manipulations. Nature 528, 358–363 (2015).
Krakauer, J. W., Ghazanfar, A. A., Gomez-Marin, A., MacIver, M. A. & Poeppel, D. Neuroscience needs behavior: correcting a reductionist bias. Neuron 93, 480–490 (2017).
Panier, T. et al. Fast functional imaging of multiple brain regions in intact zebrafish larvae using selective plane illumination microscopy. Front. Neural Circuits 7, 65 (2013).
Ahrens, M. B., Orger, M. B., Robson, D. N., Li, J. M. & Keller, P. J. Whole-brain functional imaging at cellular resolution using light-sheet microscopy. Nat. Methods 10, 413–420 (2013).
Vladimirov, N. et al. Light-sheet functional imaging in fictively behaving zebrafish. Nat. Methods 11, 883–884 (2014).
Freeman, J. et al. Mapping brain activity at scale with cluster computing. Nat. Methods 11, 941–950 (2014).
Alivisatos, A. P. et al. The brain activity map. Science 339, 1284–1285 (2013).
Klapoetke, N. C. et al. Independent optical excitation of distinct neural populations. Nat. Methods 11, 338–346 (2014).
Orger, M. B., Kampff, A. R., Severi, K. E., Bollmann, J. H. & Engert, F. Control of visually guided behavior by distinct populations of spinal projection neurons. Nat. Neurosci. 11, 327–333 (2008).
Miri, A., Daie, K., Burdine, R. D., Aksay, E. & Tank, D. W. Regression-based identification of behavior-encoding neurons during large-scale optical imaging of neural activity at cellular resolution. J. Neurophysiol. 105, 964–980 (2011).
Fang-Yen, C., Gabel, C. V., Samuel, A. D. T., Bargmann, C. I. & Avery, L. Laser microsurgery in Caenorhabditis elegans. Methods Cell Biol. 107, 177–206 (2012).
Nishimura, N. et al. Targeted insult to subsurface cortical blood vessels using ultrashort laser pulses: three models of stroke. Nat. Methods 3, 99–108 (2006).
Liu, K. S. & Fetcho, J. R. Laser ablations reveal functional relationships of segmental hindbrain neurons in zebrafish. Neuron 23, 325–335 (1999).
Mendelson, B. Development of reticulospinal neurons of the zebrafish. II. Early axonal outgrowth and cell body position. J. Comp. Neurol. 251, 172–184 (1986).
McLean, D. L. & Fetcho, J. R. Ontogeny and innervation patterns of dopaminergic, noradrenergic, and serotonergic neurons in larval zebrafish. J. Comp. Neurol. 480, 38–56 (2004).
Randlett, O. et al. Whole-brain activity mapping onto a zebrafish brain atlas. Nat. Methods 12, 1039–1046 (2015).
Thiele, T. R., Donovan, J. C. & Baier, H. Descending control of swim posture by a midbrain nucleus in zebrafish. Neuron 83, 679–691 (2014).
Roeser, T. & Baier, H. Visuomotor behaviors in larval zebrafish after GFP-guided laser ablation of the optic tectum. J. Neurosci. 23, 3726–3734 (2003).
Lemon, W. C. et al. Whole-central nervous system functional imaging in larval Drosophila. Nat. Commun. 6, 7924 (2015).
Tomer, R., Khairy, K., Amat, F. & Keller, P. J. Quantitative high-speed imaging of entire developing embryos with simultaneous multiview light-sheet microscopy. Nat. Methods 9, 755–763 (2012).
Royer, L. A. et al. Adaptive light-sheet microscopy for long-term, high-resolution imaging in living organisms. Nat. Biotechnol. 34, 1267–1278 (2016).
Dana, H. et al. Sensitive red protein calcium indicators for imaging neural activity. eLife 5, 413–420 (2016).
Kawashima, T., Zwart, M. F., Yang, C.-T., Mensh, B. D. & Ahrens, M. B. The serotonergic system tracks the outcomes of actions to mediate short-term motor learning. Cell 167, 933–946.e20 (2016).
Sugihara, I., Lang, E. J. & Llinás, R. Serotonin modulation of inferior olivary oscillations and synchronicity: a multiple-electrode study in the rat cerebellum. Eur. J. Neurosci. 7, 521–534 (1995).
Ito, M. Long-term depression as a memory process in the cerebellum. Neurosci. Res. 3, 531–539 (1986).
Bae, Y.-K. et al. Anatomy of zebrafish cerebellum and screen for mutations affecting its development. Dev. Biol. 330, 406–426 (2009).
Bouchard, M. B. et al. Swept confocally-aligned planar excitation (SCAPE) microscopy for high speed volumetric imaging of behaving organisms. Nat. Photonics 9, 113–119 (2015).
Tomer, R. et al. SPED light sheet microscopy: fast mapping of biological system structure and function. Cell 163, 1796–1806 (2015).
Quirin, S. et al. Calcium imaging of neural circuits with extended depth-of-field light-sheet microscopy. Opt. Lett. 41, 855–858 (2016).
Fahrbach, F. O., Voigt, F. F., Schmid, B., Helmchen, F. & Huisken, J. Rapid 3D light-sheet microscopy with a tunable lens. Opt. Express 21, 21010–21026 (2013).
Patterson, G. H. & Lippincott-Schwartz, J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science 297, 1873–1877 (2002).
Ahrens, M. B. et al. Brain-wide neuronal dynamics during motor adaptation in zebrafish. Nature 485, 471–477 (2012).
Motta-Mena, L. B. et al. An optogenetic gene expression system with rapid activation and deactivation kinetics. Nat. Chem. Biol. 10, 196–202 (2014).
Singh, C., Oikonomou, G. & Prober, D. A. Norepinephrine is required to promote wakefulness and for hypocretin-induced arousal in zebrafish. eLife 4, e07000 (2015).
Lovett-Barron, M. et al. Ancestral circuits for the coordinated modulation of brain state. Cell 171, 1411–1423.e17 (2017).
Severi, K. E. et al. Neural control and modulation of swimming speed in the larval zebrafish. Neuron 83, 692–707 (2014).
Wang, W.-C. & McLean, D. L. Selective responses to tonic descending commands by temporal summation in a spinal motor pool. Neuron 83, 708–721 (2014).
Horn, K. M., Deep, A. & Gibson, A. R. Progressive limb ataxia following inferior olive lesions. J. Physiol. (Lond.) 591, 5475–5489 (2013).
Masino, M. A. & Fetcho, J. R. Fictive swimming motor patterns in wild type and mutant larval zebrafish. J. Neurophysiol. 93, 3177–3188 (2005).
White, R. M. et al. Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell 2, 183–189 (2008).
Chen, T.-W. et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300 (2013).
Satou, C. et al. Transgenic tools to characterize neuronal properties of discrete populations of zebrafish neurons. Development 140, 3927–3931 (2013).
Koyama, M., Kinkhabwala, A., Satou, C., Higashijima, S. & Fetcho, J. Mapping a sensory-motor network onto a structural and functional ground plan in the hindbrain. Proc. Natl. Acad. Sci. USA 108, 1170–1175 (2011).
Preibisch, S., Saalfeld, S., Schindelin, J. & Tomancak, P. Software for bead-based registration of selective plane illumination microscopy data. Nat. Methods 7, 418–419 (2010).
Wasserstein, R. L. & Lazar, N. The ASA’s statement on p-values: context, process, and purpose. Am. Stat. 70, 129–133 (2016).
Farcomeni, A. A review of modern multiple hypothesis testing, with particular attention to the false discovery proportion. Stat. Methods Med. Res. 17, 347–388 (2008).
Zhou, W. et al. Non-sense mutations in the dihydropyridine receptor β1 gene, CACNB1, paralyze zebrafish relaxed mutants. Cell Calcium 39, 227–236 (2006).
Eaton, R. C., Lee, R. K. K. & Foreman, M. B. The Mauthner cell and other identified neurons of the brainstem escape network of fish. Prog. Neurobiol. 63, 467–485 (2001).
Murphy, D. B. & Davidson, M. W. Fundamentals of Light Microscopy and Electronic Imaging. (John Wiley & Sons, Hoboken, NJ, USA, 2012).
Zipfel, W. R., Williams, R. M. & Webb, W. W. Nonlinear magic: multiphoton microscopy in the biosciences. Nat. Biotechnol. 21, 1369–1377 (2003).
Avants, B. B. et al. The optimal template effect in hippocampus studies of diseased populations. Neuroimage 49, 2457–2466 (2010).
Acknowledgements
We thank F. Engert, N. Li, A. Bolton, A. Hantman, and B. Mensh for comments on the manuscript; V. Goncharov, D. Flickinger, C. McRaven, and Janelia Experimental Technology for help with microscopy design and construction; D. Flickinger for helpful discussions on ETL-based remote focusing; M. Zwart for discussions on optogenetics; T. Kawashima for technical assistance; M. Coleman, A. Chiu, and F. Angstadt for assistance with coding; S. Sawtelle for discussions and ETL driver construction; D. Leuenberger and C. Grätzel at Optotune for discussions; J. Cox, B. Foster, J. Rouchard, and C. McGlynn for fish husbandry; D. Bennet for assistance with Thunder code; K. Branson, S. Romani, M. Rubinov, and A. Porrello for helpful discussions; G. Ceric, K. Carlile, and R. Lines for assistance with the high-performance computer cluster; and S.-I. Higashijima (NIBB, Japan) for providing the fish lines labeling gad1b- and vGlut2a-positive neurons. This work was supported by the Howard Hughes Medical Institute (M.K., P.J.K., and M.B.A.); by the Simons Foundation–Simons Collaboration on the Global Brain Research Awards 325171 and 542943SPI (J.F. and M.B.A.); and by MDC Berlin (S.P.).
Author information
Authors and Affiliations
Contributions
N.V., CW., P.J.K., and M.B.A. conceived the project. N.V. and M.B.A. designed and built the laser ablation system for Zebrascope. N.V. created the software for brain mapping. C.W. and P.J.K. designed and built the Opto-SiMView microscope for concurrent 3D imaging and 3D optical manipulation. N.V., C.W. and B.H. created the software for brain perturbation. N.V. performed the ablation-based experiments in Figs. 1–4; C.W. and B.H. performed the optogenetics-based experiments in Figs. 5 and 6; M.T. and M.K. performed the ablation experiments in Supplementary Fig. 1c; A.P., N.V., and M.K. performed the ablation experiments in Fig. 1g,h. Y.M. contributed to cross-modal fish brain registration. J.F., J.D.W., and S.P. contributed to software. C.-T.Y. generated the Tg(elavl3:jRGECO1b) and Tg(elavl3:CoChR-eGFP) lines. P.J.K. and M.B.A. supervised the work. N.V., C.W., P.J.K. and M.B.A. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Single-cell-ablation accuracy for population ablations
a, Single-cell accuracy of two-photon laser ablation. Similar data as in Fig. 1d, but additionally showing cytosolic examples. The increase in fluorescence in cytosolic-expressing cells lasts for a shorter period than in nuclear-expressing cells. Scale bars, 10 μm. b, Electrophysiological verification of single-cell resolution of two-photon ablation. Same data as in Fig. 1f, this time including fluorescence traces from nearby cells. c, Mauthner cell ablation and control experiments (top row). The Mauthner cell was functionally identified within the reticulospinal region in Tg(elavl3:H2B-GCaMP6f) relaxed mutants (shown in red). Ablated cells are visible by their increase in brightness. (bottom row) Behavioral latency increases significantly after Mauthner cell ablation, but not after control cell ablation (means and 95% confidence intervals are shown; Mauthner ablation: p=0.027; control: p=0.345; Wilcoxon signed-rank test, asterisk indicates p<0.05). Examples of fictive behavior traces after Mauthner cell ablation and control ablations are shown. d, Single-cell resolution of ablation is preserved when ablating multiple nearby cells, both in nuclear-localized expression of the calcium indicator (left) and cytosolic expression (middle, right). Dots show the ablation coordinates, arrows are added for clarity. For both ablation types, cell brightness initially increases and eventually decreases; in the case of cytosolic expression, brightness decreases faster, visible in the example (middle and right).
Supplementary Figure 2 Two-color imaging of double-transgenic fish expressing neurotransmitter markers in red (glutamatergic vGlut2a or GABAergic gad1b) or spinal backfill, overlaid with a functional map for swimming computed from GCaMP6f imaging
a, Overlay of neurons active during swimming and a glutamatergic marker (colocalization is black, e.g. arrows).b, Overlay of neurons active during swimming and a GABAergic marker (colocalization at e.g. arrows).c, Spinal cord backfill with Texas Red dye shows that fc-RoL1 (tuned to swimming) includes RoL1 cells, and fc-nMLF includes cells of the nMLF reticulospinal group.
Supplementary Figure 3 Changes in activity of individual neurons after ablation of motor-tuned cells in fc-IO
Neurons in the optic tectum (cell 1), cerebellum (cell 2), midbrain (cells 3 and 4) significantly change their response to forward-moving stimulus after fc-IO ablation.a, Left: relative change in ∆F/F (during the forward stimulus) before versus after ablation. Right: functional map.b, Individual traces of neuron fluorescence and their averages over multiple trials (thick lines).c, Examples of dynamics of ∆F/F (top 95th-percentile of voxels in the region of interest over time) in the cerebellum, nMLF and RoL1 before and after inferior olive ablation (fish1, fish2) and random control ablations in the optic tectum (fish 3), arrows and the sizes of the circles indicate the flow of time. Experiments were done in Tg(elavl3:GCaMP6f) fish.
Supplementary Figure 4 Custom ETL driver and closed-loop optimization of ETL waveforms
a, Schematic of the custom driver for high-speed, high-precision control of the electrical tunable lens (ETL).b, Photos of the custom ETL driver.c, Plots of the voltage waveform used to operate the ETL, before (red) and after (black) closed-loop optimization (representing the control voltage time course during one full volume acquisition). The starting waveform was determined with the calibration procedure described in Methods section “Calibration procedure of the ETL-based 3D manipulation arm” and used as a starting point for closed-loop optimization. Waveform optimization during high-speed volumetric imaging was performed as described in Methods section “Closed-loop optimization of ETL control waveform”. Input voltage to the ETL driver is controlled at a sampling rate of 50 kHz, i.e. the time interval between sample steps in the plot is 20 µs. One volume scan takes 240 ms (corresponding to 12,000 samples; note that the waveforms shown here include 500 samples each preceding and succeeding the core volume scan). The focus stability obtained with this procedure during volumetric imaging is shown in Fig. 5b. d, Time-dependent correction of voltage values determined by closed-loop optimization relative to the starting waveform (i.e. difference between black and red curves in panel c).
Supplementary Figure 5 Characterization of the spatial extent of photostimulation during high-speed piezo-based volumetric imaging of photoactivateable-GFP-expressing larval zebrafisha
a, Dorsal-view maximum intensity projections and lateral-view slices of anatomy reference stacks acquired before and after photostimulation of targets in the brains of PA-GFP-expressing larval zebrafish. Targets A and B were photoactivated in the first specimen, and targets C and D were photoactivated in a second specimen. The depths of targets A, B, C, D relative to the surface of the brain were 20.5 μm, 99.5 μm, 101.5 μm and 19 μm, respectively.b, Dorsal- and lateral-view maximum intensity projections of the photoactivated regions A, B, C and D.c, Intensity profiles of photoactivated regions along lateral and axial directions and corresponding FWHM size. Overall, the spatial extent of activated regions (6-7 µm axially and 4-6 µm laterally) was found to be uniform across the brain and consistent with the resolution characterization based on a florescent dye shown in Fig. 5. The slightly smaller axial size of activated regions compared to the fluorescein-based resolution benchmark could arise from additional non-linear effects (with respect to laser power density) involved in the photoactivation of PA-GFP. The larger lateral size is the result of GFP diffusion in the cell’s cytoplasm following photoactivation. The experiment shown in this figure was repeated independently three times with similar results (for similar depths in the fish brain). Scale bars, 100 µm (a), 5 µm (b).
Supplementary Figure 6 Characterization of the spatial extent of photostimulation deep inside the larval zebrafish brain
a, Dorsal-view maximum intensity projections and lateral-view slices of anatomy reference stacks acquired before and after photostimulation of targets in the brains of PA-GFP-expressing larval zebrafish. Target E was photoactivated in specimen #3, and targets F and G were photoactivated in specimen #4 (please see SI Supplementary Fig. 5 for targets A-D in specimens #1 and #2). The depths of targets E, F and G relative to the surface of the brain were 180.0 µm, 172.5 µm and 178.0 µm, respectively.b, Dorsal- and lateral-view maximum intensity projections of the photoactivated regions E, F and G.c, Intensity profiles of photoactivated regions along lateral and axial directions and corresponding FWHM size. The spatial extent of activated regions (7-8 µm axially and 5-7 µm laterally) was found to be uniform across the brain and consistent with the resolution characterization based on a florescent dye shown in Fig. 5 (please also see the legend of SI Supplementary Fig. 5c in this context). The experiment shown in this figure was repeated independently once with similar results (for similar depths in the fish brain). Scale bars, 100 µm (a), 5 µm (b).
Supplementary Figure 7
Expression pattern of the HuC::CoChR-eGFP transgenic line a, Slices from a 3D image stack of a 5-day old transgenic zebrafish larva Tg(elval3:CoChR-eGFP) expressing the opsin CoChR (Klapoetke et al. 2014) in most neurons. The images were acquired with the Opto-SiMView light-sheet microscope.b, Enlarged views of the three regions marked by green boxes in panel a. The imaging experiment shown in this figure was repeated independently twice with similar results. Scale bars, 100 µm (a), 10 µm (b).
Supplementary Figure 8 Repetition of optogenetic excitation experiment for an additional specimen
Related to Fig. 6. Optical manipulation of dorsal raphe nucleus (a), inferior olive (b) and nucleus MLF (c) during volumetric functional imaging, using a transgenic line Tg(elavl3:jRGECO1b) x Tg(elavl3:CoChR-eGFP), as in Fig. 6. Targeted cells are indicated by yellow dots and locations of example cell responses by open circles. Shown are four maximum intensity projections, each covering a different 50 µm z-interval of the full image volume (left). The color code indicates the weighted activity change relative to control manipulations, averaged across 12 trials for targets in the DRN, 10 trials for the IO, and 6 trials for the nMLF. Responses are weighted relative to manipulations targeted to points outside the brain to de-emphasize activity responses that are not well conserved across trials and that are shared between manipulation inside and outside the brain. Control manipulations were targeted to positions outside the brain. ΔF/F time traces of the indicated positions (right) show the activity profile of selected single cells (orange, magenta, blue, green) located in the cerebellar cortex, dorsal raphe nucleus (DRN), dorsal hindbrain (DHB) and inferior olive. Traces are shown for all trials (thin colored lines) and controls (thin black lines; for these controls the two-photon laser was targeted just outside the fish’s head), as well as for their respective mean (thick lines).he experiment shown in this figure was repeated independently three times with similar results. Scale bar, 50 µm.
Supplementary Figure 9 Control optical manipulations during volumetric functional imaging in Tg(elavl3:jRGECO1b) to test for direct effects of laser exposure, shutter noise, et cetera, on neural activity
Manipulated sites are indicated by yellow dots, targeting the DRN (a), IO (b) and nMLF (c), respectively. The weighted activity change is indicated by a green-magenta diverging color map. Weighting and scaling are identical to Fig. 6. Note that the maps are generally dimmer and strong peaks are absent in comparison to Fig. 6.The experiment shown in this figure was repeated independently once with similar results (please see SI Supplementary Fig. 10 for the second example).The experiment shown in this figure was repeated independently once with similar results (please see SI Supplementary Fig. 10 for the second example).Scale bar, 50 µm.
Supplementary Figure 10 Repetition of control experiment for an additional specimen
Related to Supplementary Fig. 9. Manipulated sites are indicated by yellow dots, targeting the DRN (a), IO (b) and nMLF (c), respectively. The weighted activity change is indicated by a green-magenta diverging color map. Weighting and scaling are identical to Fig. 6. Note that the maps are generally dimmer and strong peaks are absent in comparison to Fig. 6. The experiment shown in this figure was repeated independently once with similar results (please see SI Supplementary Fig. 9 for the second example). Scale bar, 50 µm.
Supplementary Figure 11 Spatial confinement of optical manipulation of the DRN
For 9 time points (3 seconds), the sites were targeted iteratively, illuminating each site for 4 ms at a time and jumping between sites within 1 ms.a, Overlay of an anatomical reference (gray, 5th percentile F0 baseline of 159 time points, including 100 time points before manipulation and 50 time points after manipulation) and maximum-projection of ∆F/F over these 9 time points (red).b, Selected sites to quantify ∆F/F activity traces. Two manipulation sites were selected (blue). For each of these, three sites at increasing distance from the manipulation site were selected as well, in 5 µm increments (orange, green and red, respectively).c, For all sites, ∆F/F traces were extracted from pixels in a 2 µm radius (5th percentile constant baseline). Time points with manipulations are indicated with gray background. Since the manipulations occurred concurrently with imaging, the responses are not always recorded directly. For most of the sites, there are only 2 time points where the detection piezo position is exactly pointed at the targeted z-slice while the respective site is activated, resulting in two primary peaks in the respective ∆F/F traces. The experiment shown in this figure was repeated independently three times with similar results. Scale bar, 25 µm.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–11, Supplementary Tables 1–3 and Supplementary Note
Supplementary Video 1
Three-dimensional renderings of the zebrafish brain functional map shown in Fig. 1c, in a fish with nuclear-localized GCaMP6f expression. Colors indicate neuronal response tuning to swimming (red), forward stimulus (grayscale) and backward stimulus (blue). Functional maps with similar features in terms of the most-active brain areas were obtained for more than 50 fish
Supplementary Video 2
Functional map of a cytosolic-expressing GCAMP6f fish, shown at different z-planes. Maximum-intensity projections of this functional map are shown in Fig. 2a. Functional maps with similar features in terms of the most-active brain areas were obtained for more than 30 fish
Supplementary Video 3
Ablation of single neurons during light-sheet imaging of the same plane: The fish performed the optomotor response while a single plane in its hindbrain was imaged at 50 Hz and four individual cells were ablated. These are the same cells as shown in Fig. 1d (top) and Suppl. Fig. 1a (left). The cells become very bright and lose correlation with stimulus and behavior immediately after ablation. Time is shown relative to the moment of laser exposure (t = 0). Single-plane high-speed imaging and ablation experiments were carried out in 12 fish with similar results
Supplementary Video 4
Three-dimensional renderings of the brain-wide ablation effects in Fig. 2, overlaid on reference anatomy brain (Z-Brain) in gray: Differences in neuronal activity ΔF/F, averaged over the forward-stimulus period, before and after targeted ablations. Green indicates a decrease in activity after ablation. The numbers of fish brains used in generating each map are provided in Fig. 2c-h
Supplementary Data 1
Zemax model of the ETL-based optical manipulation arm for 3D targeted two-photon manipulation during volumetric light-sheet imaging
Supplementary Data 2
Parts list and PCB schematics of the custom driver for Optotune ETLs
Rights and permissions
About this article
Cite this article
Vladimirov, N., Wang, C., Höckendorf, B. et al. Brain-wide circuit interrogation at the cellular level guided by online analysis of neuronal function. Nat Methods 15, 1117–1125 (2018). https://doi.org/10.1038/s41592-018-0221-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-018-0221-x
This article is cited by
-
The influence of cortical activity on perception depends on behavioral state and sensory context
Nature Communications (2024)
-
Automatic monitoring of neural activity with single-cell resolution in behaving Hydra
Scientific Reports (2024)
-
Real-time analysis of large-scale neuronal imaging enables closed-loop investigation of neural dynamics
Nature Neuroscience (2024)
-
FIOLA: an accelerated pipeline for fluorescence imaging online analysis
Nature Methods (2023)
-
A single-cell transcriptomic atlas of complete insect nervous systems across multiple life stages
Neural Development (2022)