Abstract
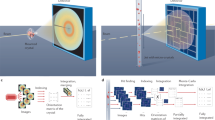
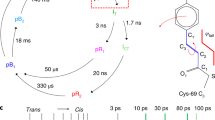
We present a ‘hit-and-return’ (HARE) method for time-resolved serial synchrotron crystallography with time resolution from milliseconds to seconds or longer. Timing delays are set mechanically, using the regular pattern in fixed-target crystallography chips and a translation stage system. Optical pump-probe experiments to capture intermediate structures of fluoroacetate dehalogenase binding to its ligand demonstrated that data can be collected at short (30 ms), medium (752 ms) and long (2,052 ms) intervals.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Bar-Even, A. et al. Biochemistry 50, 4402–4410 (2011).
Fenwick, R. B., Esteban-Martín, S. & Salvatella, X. Eur. Biophys. J. 40, 1339–1355 (2011).
Gati, C. et al. IUCrJ 1, 87–94 (2014).
Standfuss, J. & Spence, J. IUCrJ 4, 100–101 (2017).
Stellato, F. et al. IUCrJ 1, 204–212 (2014).
Weinert, T. et al. Nat. Commun. 8, 542 (2017).
Oghbaey, S. et al. Acta Crystallogr. D Struct. Biol. 72, 944–955 (2016).
Sherrell, D. A. et al. J. Synchrotron Radiat. 22, 1372–1378 (2015).
Anfinrud, P. A., Lim, M. & Jackson, T. A. Prog. Biophys. Mol. Biol. 65, S4 (1996).
Schotte, F. et al. Science 300, 1944–1947 (2003).
Kubo, M. et al. J. Synchrotron Radiat. 24, 1086–1091 (2017).
Chan, P. W., Yakunin, A. F., Edwards, E. A. & Pai, E. F. J. Am. Chem. Soc. 133, 7461–7468 (2011).
Kim, T. H. et al. Science 355, eaag2355 (2017).
Givens, R. S. et al. Can. J. Chem. 89, 364–384 (2011).
Levantino, M., Yorke, B. A., Monteiro, D. C. F., Cammarata, M. & Pearson, A. R. Curr. Opin. Struct. Biol. 35, 41–48 (2015).
Spence, J. C. H. Faraday Discuss. 171, 429–438 (2014).
Spence, J. C. H. IUCrJ 4, 322–339 (2017).
Chavas, L. M., Gumprecht, L. & Chapman, H. N. Struct. Dyn. 2, 041709 (2015).
Olmos, J. L. Jr. et al. BMC Biol. 16, 59 (2018).
Beyerlein, K. R. et al. IUCrJ 4, 769–777 (2017).
Owen, R. L. et al. Acta Crystallogr. D Struct. Biol. 73, 373–378 (2017).
Schulz, E. C. et al. J. Appl. Crystallogr. 50, 1773–1781 (2017).
Mueller, C. et al. Struct. Dyn. 2, 054302 (2015).
Zarrine-Afsar, A. et al. Acta Crystallogr. D Biol. Crystallogr. 68, 321–323 (2012).
Epp, S. W. et al. Struct. Dyn. 4, 054308 (2017).
Muller-Werkmeister, H. M. et al. Biophys. J. 110, 513a (2016).
Kabsch, W. Acta Crystallogr. D Biol. Crystallogr. 70, 2204–2216 (2014).
McCoy, A. J. et al. J. Appl. Crystallogr. 40, 658–674 (2007).
Adams, P. D. et al. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Painter, J. & Merritt, E. A. Acta Crystallogr. D Biol. Crystallogr. 62, 439–450 (2006).
Liebschner, D. et al. Acta Crystallogr. D Struct. Biol. 73, 148–157 (2017).
Lang, P. T., Holton, J. M., Fraser, J. S. & Alber, T. Proc. Natl. Acad. Sci. USA 111, 237–242 (2014).
Acknowledgements
Synchrotron MX data were collected at beamline P14 operated by EMBL Hamburg at the PETRA III storage ring (DESY, Hamburg, Germany). The authors thank T. Schneider and G. Bourenkov for exceptional beamline support and, along with A. Pearson, discussions about the experiment. The authors are grateful to W. Kabsch and K. Diederichs for outstanding support in data processing and helpful discussions. Parts of this research were also carried out at beamline P11 at PETRA III at DESY, a member of the Helmholtz Association (HGF). We gratefully recognize A. Burkhardt and O. Lorbeer for assistance in using P11. We thank L. Andricek of the Max Planck Semiconductor lab for fabrication of the crystallography chips. We gratefully acknowledge the support provided by the Max Planck Society and the excellence cluster “The Hamburg Centre for Ultrafast Imaging—Structure, Dynamics and Control of Matter at the Atomic Scale” of the Deutsche Forschungsgemeinschaft (R.J.D.M.) and the People Programme (Marie Curie Actions) of the European Union’s Seventh Framework Programme (FP7/2007–2013) under REA grant agreement no. 623994 (H.M.M.-W.). P.M. is the recipient of an Alexander von Humboldt-Stiftung postdoctoral fellowship. Work at the ETH was generously supported by the ETH Research Council (ETH-01 13-2) (F.D.). E.F.P. gratefully acknowledges funding from the Natural Sciences and Engineering Research Council of Canada (award no. RGPIN-2015- 04877), the Canada Research Chairs program and the Burroughs Wellcome Fund for a Collaborative Research Travel Grant.
Author information
Authors and Affiliations
Contributions
E.C.S., P.M., H.M.M.-W., E.F.P. and R.J.D.M. designed the experiment. E.C.S., P.M. and H.M.M.-W. conducted the experiments with support from F.T., E.F.P. and W.S. E.P., R.D.G. and F.D. designed a reaction scheme and synthesized the caged compound. P.M. prepared the protein crystals. F.T. designed the experimental endstation and, together with P.M., developed the HARE algorithms. H.M.M.-W. and A.J. built the optics part of the setup. E.C.S. and P.M. analyzed and processed the diffraction data and wrote the manuscript. All authors discussed and corrected the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Homodimeric fluoroacetate dehalogenase and photolytic generation of its substrate fluoroacetate.
a, The photocaged substrate for FAcD pHP-FAc is sensitive to photolysis initiated by UV light pulses with a wavelength between 320 nm and 360 nm. Cleavage of the phenol group releases FAcD’s substrate fluroacetate. b, A cartoon representation of homodimeric FAcD with the backbone of subunit A in yellow and subunit B in orange. The location of the cap domain (cyan) and the positions of the active site residues (purple) are indicated.
Supplementary Figure 2 End station design.
a, Top-down schematic of a typical light-triggered SSX arrangement. The chip can be viewed from an IR camera using a mirror mounted on a capillary which is illuminated via a retractable IR backlight. Alignment and chip movement are done via the translation stage system. Samples on the chip are pumped via a femtosecond laser and probed via the X-ray beam; the diffraction cone is represented in green. b, CAD representation of a. c, A CAD representation of the end station with and without the detector. The diffraction cone is represented in green.
Supplementary Figure 3 The HARE approach with chip gen. 1.
A representation of a single compartment on the chip with 12 × 12 features. X-ray pulses are depicted in green, and laser pulses are shown in blue. In order to have a large number of valid HARE times, it is most efficient for the number of features per compartment row and column to be a highly composite number (e.g., 12). Longer times than a HARE value of 12 would therefore need to be certain multiples of 12 (e.g., 24, 36, 48, etc.). Data collection for an HARE of 24 would be as follows: Starting from row a, the first feature in the first column (aa) is exposed to an X-ray pulse and a subsequent dark image is recorded. Immediately following the X-ray pulse the sample is pumped with the femtosecond laser for t = 0. This procedure is repeated for a total of 24 features. After pumping of the 24 features, the stages move back to their starting positions. The same 24 features are then re-probed with X-rays after a delay time matching the total time for probe-pumping of the 24 features. For every probed feature on the chip, the resulting data include both a dark image and an exposed image with a corresponding delay dependent on the HARE number.
Supplementary Figure 4 Pulse sequence.
The pulse sequence plot shows the relative on and off times of the various instruments of our setup. Initially a dark image is taken: (1) the stage moves into position, (2) the detector is set to exposure mode, and (3) after a dark image is acquired a TTL pulse triggers the laser. After the laser pulse there is a delay corresponding to the HARE number, during which further images are acquired. After the delay the stages move back to their original position and the sequence is repeated without laser excitation.
Supplementary Figure 5 Description of the HARE formalism.
Let the coordinates (v, w) denote the index of the compartment in the horizontal and vertical directions, and let (x, y) be the index of the feature inside the compartment. With Nv and Nw as the number of compartments in the horizontal and vertical directions, Nx and Ny as the number of features per compartment in the horizontal and vertical directions, and C as the HARE constant, the coordinates can be determined by \(v=\frac{i-1}{2{N}_{f}}\) and Nv+1 \(w=\frac{i-1}{2{N}_{v}{N}_{f}}\) +1 \(x{\rm{=}}\frac{j}{2C}C\) mod Nx+j mod min (Nx, C)+1 \(y=\frac{j}{2C}\frac{C}{{N}_{x}}\) mod Nx+\(\frac{j}{{N}_{x}}\) mod \(\frac{C}{{N}_{x}}\) +1 with x as the floor function, Nc = NvNw, Nf = NxNy, j = (i – 1) mod 2Nf and i as the exposure index ranging from 1 to the total number of exposures 2NcNf. These four equations apply if C|Nx for C ≤ Nx or if Nx|C ∧ C|Nf otherwise. Laser excitation takes place if (i – 1) mod 2C < C. In order to have a high number of valid HARE times, it is helpful to choose Nx and Ny as highly composite numbers. The number of possible HARE times can be calculated by σ0(Nx) + σ0(Ny) – 1, where σx(n) is the divisor function.
Supplementary Figure 6 Infrared images of the chip.
a, An empty chip is shown as a control. The yellow arrows point out underdeveloped features, which appear semi-transparent in the IR light. b, Loaded chip with yellow arrows representing crystals sitting in the features of the chip. Crystals sitting inside of the features can be clearly distinguished from crystals on its surface and from empty features.
Supplementary Figure 7 Cumulative distribution function for time delays.
The plot represents the deviation in milliseconds for five representative HARE values (2, 3, 12, 72 and 144) from their respective median values. The individual median values for each of the five HARE times are set to 0 ms for comparison in the plot. Although the greatest HARE values have the largest deviation from the median, the total percent difference is comparatively small. Data were derived from the total features of a randomly selected chip per time point.
Supplementary Figure 8 Absolute value END-RAPID maps.
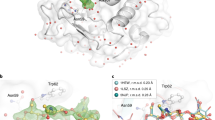
FAcD electron density changes as a function of time. All electron density maps are represented as absolute value END-RAPID maps at a value of 0.3e A–3 σ. The electron density for the bound ligand in active site A is highlighted by a red circle. The flexible side chain of Tyr149 is indicated by a red arrow. The absolute value maps allow for quantitative comparability of the structures by placing all the different time points on an absolute scale, thereby emphasizing that the reduction of the electron observed in the cap domain is a result of enzymatic turnover and not an artifact originating from σ offset of the maps. However, the observed differences recapitulate the changes in the electron density seen in the POLDER-OMIT maps.
Supplementary Figure 9 Fobs–Fobs difference density maps.
Difference electron density maps, shown in green (positive) and red (negative), of the 30-ms, 752-ms and 2,052-ms structures compared with the 0-ms structure contoured at 3.0σ. There are few to no differences in the active sites of the 30-ms structure. There is a large influx of density in subunit A of the 752-ms structure, which is indicative of the Michaelis complex. Differences in the active site in the 2,052-ms structure are readily observed only when the contour levels are decreased to 2.0σ. This is probably due to insufficient signal to noise. Whereas the cap domain in subunit A remains constant over time points, the cap domain in subunit B shows a consistent trend of increased dynamics as indicated by the increase in negative density as the reaction proceeds.
Supplementary Figure 10 Radiation damage.
The figure shows absolute value END-RAPID (a) and POLDER-OMIT (b) maps for a structure exposed to 0.28 MGy and 0.56 Mgy, respectively. The maps clearly show equivalent electron density for the active site and the cap-domain loop in both subunits, irrespective of the cumulative radiation dose. This not only shows the radiation insensitivity of the enzyme but also demonstrates that the disordering of the cap-domain loop is associated with enzymatic turnover and not induced by radiation damage.
Supplementary Figure 11 Structure comparison.
The figure shows cartoon representations of FAcD with the 0-ms structure in yellow, the previously solved structure PDB 5K3C in purple and the 2,052-ms structure in green. The superpositioning reveals minimal r.m.s. deviation differences between the respective time-point structures and the previously solved cryo-structure (a, b) and even smaller differences between the two time-point structures (c).
Supplementary information
Supplementary Text and Figures
Supplementary Figs. 1–11, Supplementary Results, Supplementary Discussion, Supplementary Tables 1 and 2, and Supplementary Note
Rights and permissions
About this article
Cite this article
Schulz, E.C., Mehrabi, P., Müller-Werkmeister, H.M. et al. The hit-and-return system enables efficient time-resolved serial synchrotron crystallography. Nat Methods 15, 901–904 (2018). https://doi.org/10.1038/s41592-018-0180-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-018-0180-2
This article is cited by
-
Millisecond cryo-trapping by the spitrobot crystal plunger simplifies time-resolved crystallography
Nature Communications (2023)
-
Serial femtosecond crystallography
Nature Reviews Methods Primers (2022)
-
Serial protein crystallography in an electron microscope
Nature Communications (2020)
-
Liquid application method for time-resolved analyses by serial synchrotron crystallography
Nature Methods (2019)