Abstract
Anti-CRISPR proteins are powerful tools for CRISPR–Cas9 regulation; the ability to precisely modulate their activity could facilitate spatiotemporally confined genome perturbations and uncover fundamental aspects of CRISPR biology. We engineered optogenetic anti-CRISPR variants comprising hybrids of AcrIIA4, a potent Streptococcus pyogenes Cas9 inhibitor, and the LOV2 photosensor from Avena sativa. Coexpression of these proteins with CRISPR–Cas9 effectors enabled light-mediated genome and epigenome editing, and revealed rapid Cas9 genome targeting in human cells.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
Vectors will be made available via Addgene (#113033–113042 and #113129–113133) and upon request. Annotated vector sequences (GenBank files), the KNIME image analysis pipeline and PDB files of the CASANOVA model are provided as Supplementary Data. Code and data for the computational domain assembly and the design of improved Acr–LOV mutants are available on GitHub (https://github.com/zanderharteveld/anti-CRISPR-designs), and source data for Figs. 1 and 2 are available online.
References
Barrangou, R. & Doudna, J. A. Nat. Biotechnol. 34, 933–941 (2016).
Richter, F. et al. Curr. Opin. Biotechnol. 48, 119–126 (2017).
Rauch, B. J. et al. Cell 168, 150–158 (2017).
Hynes, A. P. et al. Nat. Microbiol. 2, 1374–1380 (2017).
Basgall, E. M. et al. Microbiology 164, 464–474 (2018).
Shin, J. et al. Sci. Adv. 3, e1701620 (2017).
Harper, S. M., Neil, L. C. & Gardner, K. H. Science 301, 1541–1544 (2003).
Hoffmann, M. D., Bubeck, F., Eils, R. & Niopek, D. Adv. Biosyst. 2, 1800098 (2018).
Dagliyan, O. et al. Science 354, 1441–1444 (2016).
Kim, I. et al. Sci. Rep. 8, 3883 (2018).
Dong, D. et al. Nature 546, 436–439 (2017).
Yang, H. & Patel, D. J. Mol. Cell 67, 117–127 (2017).
Brinkman, E. K., Chen, T., Amendola, M. & van Steensel, B. Nucleic Acids Res. 42, e168 (2014).
Strickland, D. et al. Nat. Methods 9, 379–384 (2012).
Strickland, D. et al. Nat. Methods 7, 623–626 (2010).
Hu, J. H. et al. Nature 556, 57–63 (2018).
Hilton, I. B. et al. Nat. Biotechnol. 33, 510–517 (2015).
Chen, B. et al. Cell 155, 1479–1491 (2013).
Pawluk, A. et al. Cell 167, 1829–1838 (2016).
Vehlow, C. et al. Bioinformatics 27, 1573–1574 (2011).
Kabsch, W. & Sander, C. Biopolymers 22, 2577–2637 (1983).
Huang, P. S. et al. PLoS One 6, e24109 (2011).
Canutescu, A. A. & Dunbrack, R. L. Jr. Protein Sci. 12, 963–972 (2003).
Coutsias, E. A., Seok, C., Jacobson, M. P. & Dill, K. A. J. Comput. Chem. 25, 510–528 (2004).
Mandell, D. J., Coutsias, E. A. & Kortemme, T. Nat. Methods 6, 551–552 (2009).
Fleishman, S. J. et al. PLoS One 6, e20161 (2011).
Engler, C., Kandzia, R. & Marillonnet, S. PLoS One 3, e3647 (2008).
Senís, E. et al. Biotechnol. J. 9, 1402–1412 (2014).
Börner, K. et al. Nucleic Acids Res. 41, e199 (2013).
Livak, K. J. & Schmittgen, T. D. Methods 25, 402–408 (2001).
Chu, V. T. et al. Nat. Biotechnol. 33, 543–548 (2015).
Acknowledgements
This study was funded by the Helmholtz association, the German Research Council (DFG) and the Federal Ministry of Education and Research (BMBF) (to R.E.). We thank M. Gunkel (BioQuant, Heidelberg) and the German Network for Bioinformatics Infrastructure (HD.HuB as part of de.NBI; grant 031A537C) for support with image analysis in KNIME. We thank K. Rippe (German Cancer Research Center (DKFZ), Heidelberg, Germany) and E. Wiedtke (Heidelberg University Clinics, Heidelberg, Germany) for providing cell lines, F. Zhang (Broad Institute of MIT and Harvard, Cambridge, MA, USA) for providing vector pSpCas9(BB)-2A-GFP, C. Gersbach (Department of Biomedical Engineering and Center for Genomic & Computational Biology, Duke University, Durham, NC, USA) for the dCas9–p300 construct, E. Sontheimer (RNA Therapeutics Institute, University of Massachusetts Medical School, Worcester, MA, USA) for pEJS477-pHAGE-TO-SpydCas9_3XmCherry-sgRNA/Telomere-All-in-one, the Synthetic Biology group (Heidelberg University) and Intelligent Imaging group (DKFZ, Heidelberg) for helpful discussions, and K. Niopek for critical reading of the manuscript. This work was supported by the Helmholtz International Graduate School for Cancer Research (DKFZ, Heidelberg; scholarship to M.D.H.), the German Center for Infection Research (DZIF; TTU-HIV 04.803 to K.B. and D.G.), the Cystic Fibrosis Foundation Therapeutics (CFFT; grant GRIMM15XX0 to J.F. and D.G.), Transregional Collaborative Research Center TRR179 (DFG; to C.S., D.G. and R.E.), the Cluster of Excellence CellNetworks (DFG; EXC81 to J.F., C.S. and D.G.), the European Research Council (starting grant 716058 to B.E.C.), the Swiss National Science Foundation (B.E.C. and Z.H.) and the Biltema Foundation (B.E.C.). The computational simulations were performed at the CSCS—Swiss National Supercomputing Centre through a grant obtained by B.E.C. Part of the computational work was supported by EPFL through the use of the facilities of its Scientific IT and Application Support Center.
Author information
Authors and Affiliations
Contributions
Z.H. and S.A. contributed equally to this work. F.B. and D.N. conceived the study. F.B., M.D.H., A.B., M.C.W. and D.N. designed experiments. F.B., M.D.H., S.A. and A.B. conducted experiments. F.B., M.D.H. and D.N. analyzed and interpreted data. Z.H. and B.E.C. performed computational analysis and structural modeling. J.F. developed the luciferase cleavage reporter construct and the CFTR-targeting gRNA. C.S. and L.D. made practical contributions. K.B. and D.G. provided expertise and practical support for AAV experiments. R.E. and D.N. jointly directed the work. D.N. wrote the manuscript with support from all other authors.
Corresponding authors
Ethics declarations
Competing interests
F.B., M.D.H., A.B., M.C.W., J.F., L.D., D.G., R.E. and D.N. have filed European patent applications (17196813.4) related to this work.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
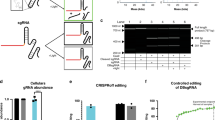
Supplementary Figure 1 Sampling of LOV2 insertion sites in AcrIIA4 loop 5.
(a) Acr–LOV hybrid generation. The Avena sativa LOV2 domain coding sequence from phototropin-1 was inserted at different positions in AcrIIA4 loop 5. NLS, SV40 nuclear localization signal. CMV, cytomegalovirus promoter. (b) Luciferase reporter cleavage assay measuring Cas9 inhibition by different Acr–LOV hybrids. HEK293T cells were co-transfected with vectors encoding (i) the Acr–LOV hybrid, (ii) Cas9 and (iii) a luciferase reporter as well as a gRNA targeting the luciferase gene. Luciferase activity was assessed 48 h post-transfection. The AcrIIA4 residue behind which the LOV2 domain was inserted is indicated. Box plots show the median (center line), first and third quartiles (box edges), 1.5× the interquartile range (whiskers) and individual data points (circles). n = 3 biologically independent samples (cell cultures). Wt, wild-type. Acr-2A-LOV2, control construct co-expressing wild-type AcrIIA4 and the LOV2 domain via a P2A sequence. ***P = 0.0004 (no Acr), ***P = 3.63 × 10–5 (wt Acr) and ***P = 8.87 × 10–5 (Acr-P2A-LOV2) by two-sided Student’s t-test.
Supplementary Figure 2 Schematic and sequences of Acr–LOV hybrids.
NLS, SV40 nuclear localization signal.
Supplementary Figure 3 Cas9 inhibition is dose dependent.
HEK293T cells were co-transfected with plasmids encoding (i) Acr–LOV hybrid, (ii) Cas9 and (iii) a luciferase reporter as well as a gRNA targeting the luciferase gene. The vector mass ratio of the transfected Cas9 and Acr–LOV construct was varied between 10:1 and 1:1, as indicated. Six hours post-transfection, cells were irradiated with pulsatile blue light (5 s ON, 10 s OFF; 2.5 W per m2) for 30 h or kept in the dark as control before assessing luciferase activity. Box plots show the median (center line) and first and third quartiles (box edges), 1.5× the interquartile range (whiskers) and individual data points (circles). n = 9 biologically independent samples (cell cultures) for the Acr–LOV hybrid 8 (1:1) and n = 12 biologically independent samples (cell cultures) for all other conditions.
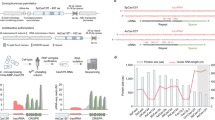
Supplementary Figure 4 CASANOVA computational model.
Structural model of CASANOVA bound to a Cas9–gRNA complex (left). The three most populated clusters of CASANOVA conformations obtained through domain assembly simulations (Methods) are displayed on the right.
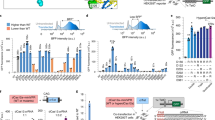
Supplementary Figure 5 Photoactivatable genome editing with CASANOVA.
Light-mediated indel mutation of human CFTR locus (a,b; with two different gRNAs), mir-122 locus (c) and VEGFA locus (d). HEK293T cells were co-transduced with AAV vectors encoding CASANOVA, Cas9 and the indicated gRNA and exposed to blue light for 70 h or kept in the dark as control. The target locus was then PCR amplified with primers flanking the estimated break point and the amplicon was denatured and re-annealed in a thermocycler to allow heteroduplex formation. Following digestion with T7 endonuclease, samples were analyzed on a 2% agarose gel (see Methods for details). Representative gel images and corresponding quantifications of editing frequencies are shown. Wt, wild-type. Box plots show the median (center line) and first and third quartiles (box edges), 1.5× the interquartile range (whiskers) and individual data points (circles). n = 3 independent experiments. All statistics by two-sided Student’s t-test. (a) ***P = 5.83 × 10–5 (b) **P = 0.0039 (c) ***P = 2.85 × 10–5 (d) ***P = 2.01 × 10–5.
Supplementary Figure 6 TIDE sequencing analysis of light-mediated indel mutation.
HEK293T cells were co-transduced with AAV vectors expressing the Cas9, CASANOVA and a gRNA targeting the CCR5 (a) or EMX1 (b) locus. Cells were exposed to blue light for 70 h or kept in the dark as control, followed by TIDE sequencing1. The target locus was PCR-amplified with primers flanking the expected breakpoint followed by Sanger sequencing of the amplicon. Total editing efficiencies and frequencies of individual insertions or deletions were then calculated by decomposition of the sequencing chromatogram using the TIDE web tool (https://www.deskgen.com/landing/tide.html). TIDE sequencing revealed a broad range of different insertions and deletions in the light, but not in the dark control samples. Data represent a single experiment.
Supplementary Figure 7 Cas9 inhibition can be modulated via mutations that affect docking of the LOV2 terminal helices.
(a) Light-dependent luciferase reporter cleavage mediated by different Acr–LOV hybrid mutants. HEK293T cells were co-transfected with plasmids encoding (i) the indicated Acr–-LOV hybrid variant, (ii) Cas9 and (iii) a luciferase reporter as well as a gRNA targeting the luciferase gene. Six hours post-transfection, cells were irradiated with pulsatile blue light for 48 h or kept in the dark as control before assessing luciferase activity. Box plots show the median (center line) and first and third quartiles (box edges), 1.5× the interquartile range (whiskers) and individual data points (circles). n = 3 biologically independent samples (cell cultures). (b) T7 endonuclease assays and (c) corresponding quantification of light-mediated indel mutation of the human CCR5 locus. HEK293T cells were co-transfected with constructs expressing the Cas9, the CCR5-locus-targeting gRNA and the indicated Acr–LOV hybrid variant. During transfection, the vector mass ratio of Acr–LOV:Cas9 construct was varied as indicated. Subsequently, cells were exposed to blue light for 70 h or kept in the dark as control. The target locus was then PCR-amplified with primers flanking the estimated break point and the amplicon was denatured and re-annealed in a thermocycler to allow heteroduplex formation. Following digestion with T7 endonuclease, samples were analyzed on a 2% agarose gel (see Methods for details). (c) Data are means and dots indicate individual data points. n = 2 independent experiments.
Supplementary Figure 8 Acr–LOV hybrids carrying a C450A LOV2 pseudodark mutation are still light responsive.
(a) Light-dependent luciferase reporter cleavage mediated by different Acr–LOV hybrid pseudodark mutants. HEK293T cells were co-transfected with plasmids encoding (i) the indicated Acr–LOV hybrid variant, (ii) Cas9 and (iii) a luciferase reporter as well as a gRNA targeting the luciferase gene. Six hours post-transfection, cells were irradiated with pulsatile blue light for 48 h or kept in the dark as control before assessing luciferase activity. Box plots show the median (center line) and first and third quartiles (box edges), 1.5× the interquartile range (whiskers) and individual data points (circles). n = 6 biologically independent samples (cell cultures). (b) T7 endonuclease assays and (c) corresponding quantification of light-mediated indel mutation of the human CCR5 locus. HEK293T cells were co-transfected with constructs expressing Cas9, CCR5-locus-targeting gRNA and the indicated Acr–LOV hybrid variant and exposed to blue light for 70 h or kept in the dark as control. During transfection, the vector mass ratio of Acr–LOV:Cas9 construct was varied as indicated. n.d., not determined. (b,c) Data correspond to a single experiment.
Supplementary Figure 9 In silico docking analysis reveals Acr mutations that improve CASANOVA performance.
(a) Light-dependent luciferase reporter cleavage mediated by different Acr–LOV hybrid mutants. HEK293T cells were co-transfected with vectors encoding (i) the indicated Acr–LOV hybrid variant, (ii) Cas9 and (iii) a luciferase reporter as well as a gRNA targeting the luciferase gene. Six hours post-transfection, cells were irradiated with pulsatile blue light for 48 h or kept in the dark as control before assessing the luciferase activity. Box plots show the median (center line) and first and third quartiles (box edges), 1.5× the interquartile range (whiskers) and individual data points (circles). n = 3 biologically independent samples (cell cultures). (b) T7 endonuclease assays and (c) corresponding quantification of light-mediated indel mutation of the human CCR5 locus. HEK293T cells were co-transfected with constructs expressing Cas9, the CCR5-locus-targeting gRNA and the indicated Acr–LOV hybrid variant and exposed to blue light for 70 h or kept in the dark as control. During transfection, the vector mass ratio of Acr–LOV:Cas9 construct was varied as indicated. Data represent a single experiment. n.d., not determined.
Supplementary Figure 10 Comparison of light-dependent indel mutation by CASANOVA and its corresponding S46D and T16F mutants.
HEK293T cells were co-transfected with constructs expressing Cas9, a gRNA and the indicated CASANOVA variant and exposed to blue light for 70 h or kept in the dark as control. During transfection, the vector mass ratio of Acr–LOV:Cas9 construct was varied as indicated. Editing frequencies were evaluated by mismatch-sensitive T7 endonuclease assay. (a) Indel mutation of CCR5 locus and (b) indel mutation of EMX1 locus. (a-b) Box plots show the median (center line) and first and third quartiles (box edges), 1.5× the interquartile range (whiskers) and individual data points (circles). n = 4 independent experiments.
Supplementary Figure 11 Optogenetic control of xCas9.
Light-dependent luciferase reporter cleavage mediated by different Acr–LOV hybrid mutants. HEK293T cells were co-transfected with vectors encoding (i) the indicated Acr–LOV hybrid variant, (ii) xCas9 and (iii) a luciferase reporter as well as a gRNA targeting the luciferase gene. Six hours post-transfection, cells were irradiated with pulsatile blue light for 48 h or kept in the dark as control before assessing luciferase activity. Box plots show the median (center line) and first and third quartiles (box edges), 1.5× the interquartile range (whiskers) and individual data points (circles). n = 6 biologically independent samples (cell cultures) for CASANOVA (S46D) and n = 9 biologically independent samples (cell cultures) for all other conditions.
Supplementary Figure 12 CASANOVA-mediated IL1RN gene activation is reversible.
Light-dependent IL1RN gene activation in HEK293T cells expressing CASANOVA and a dCas9–p300 fusion targeted to the IL1RN promoter via a combination of four gRNAs (triangles) or the gRNA mix only (round dots; control). Cells were exposed to blue light or kept in the dark as indicated in the figure, and IL1RN expression was assessed by quantitative RT-PCR at the indicated time points. Gene activation in the light-induced CASANOVA sample (day 2) continues to rise when prolonging illumination, but decreases upon withdrawal of the light stimulus. Data are means ± s.e.m. n = 7 biologically independent samples (cell cultures).
Supplementary Figure 13 Fluorescence signal of single labeled telomeres increased after CASANOVA activation.
We calculated telomere fluorescence by multiplying the area of the fluorescent spot with its mean fluorescence intensity. Box plots show the median (center line) and first and third quartiles (box edges), 1.5× the interquartile range (whiskers) and individual data points (dots). Data correspond to the experiment described in Fig. 2d,e. The total number of telomeres analyzed is indicated. ***P < 2.2 × 10–16 by a two-sided Wilcoxon rank-sum test.
Supplementary Figure 14 Reversible recruitment of Acr–LOV hybrid 5 to a plasma-membrane-targeted dCas9–gRNA complex.
(a) Light-inducible recruitment of a LOV–Acr hybrid fused to mCherry to a plasma-membrane-targeted dCas9–mVenus. (b) Representative fluorescence images of HEK293T cells expressing LOV–Acr hybrid 5 fused to mCherry and an mVenus–dCas9 fusion targeted to the plasma membrane. Cells were irradiated with blue light pulses every 30 s using a 488-nm laser for 20 min, followed by 20 min dark recovery. mVenus and mCherry fluorescence images were recorded every 5 min. Dashed lines indicate the nucleus boundary. Yellow boxes in the Acr–LOV–mCherry images correspond to plasma membrane close-up views shown below the images. Scale bar, 20 µm. (c) Quantification of the plasma membrane to cytoplasmic mCherry fluorescence ratio over time. Data are means ± s.e.m. n = 4 cells from biologically independent samples (cell cultures).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–14, Supplementary Tables 1–4, Supplementary Notes 1–7 and Supplementary Discussion
Supplementary Protocol
CASANOVA step by step: a protocol for optogenetic control of CRISPR–Cas9 with an engineered light-dependent anti-CRISPR protein
Supplementary Data
Annotated vector sequences (GenBank files), KNIME image-analysis pipeline and PDB files of the CASANOVA model
Source data
Rights and permissions
About this article
Cite this article
Bubeck, F., Hoffmann, M.D., Harteveld, Z. et al. Engineered anti-CRISPR proteins for optogenetic control of CRISPR–Cas9. Nat Methods 15, 924–927 (2018). https://doi.org/10.1038/s41592-018-0178-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-018-0178-9
This article is cited by
-
Spatiotemporal control of RNA metabolism and CRISPR–Cas functions using engineered photoswitchable RNA-binding proteins
Nature Protocols (2024)
-
Optimization of Cas9 activity through the addition of cytosine extensions to single-guide RNAs
Nature Biomedical Engineering (2023)
-
Light-switchable transcription factors obtained by direct screening in mammalian cells
Nature Communications (2023)
-
Anti-CRISPR proteins: a weapon of phage-bacterial arm race for genome editing
The Nucleus (2023)
-
Domäneninsertion: Untersuchung zur Konstruktion schaltbarer Hybridproteine
BIOspektrum (2023)